2ZL0
 
 | | Crystal structure of H.pylori ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2008-04-02 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
2ZL4
 
 | |
2ZL2
 
 | | Crystal structure of H.pylori ClpP in complex with the peptide NVLGFTQ | | Descriptor: | A peptide substrate-NVLGFTQ, A peptide substrate-NVLGFTQ for Chain R and S, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2008-04-02 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
6HWN
 
 | | Structure of Thermus thermophilus ClpP in complex with a tripeptide. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, Unknown tripeptide | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
2ZL3
 
 | | Crystal structure of H.pylori ClpP S99A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2008-04-02 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
8XOP
 
 | |
8XN4
 
 | |
4JCR
 
 | | ClpP1 N165D mutant from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zeiler, E, List, A, Alte, F, Gersch, M, Wachtel, R, Groll, M, Sieber, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2DEO
 
 | |
7WID
 
 | | Crystal structure of Staphylococcus aureus ClpP in complex with ZG180 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2022-01-03 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7WGS
 
 | | Structure of ClpP from Staphylococcus aureus in complex with (S)-ZG197 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2R)-butan-2-yl]-8-[(1S)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7WH5
 
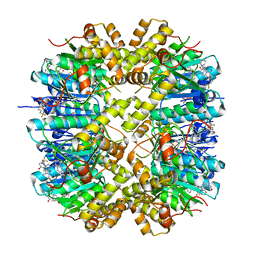 | | Crystal structure of human ClpP in complex with ZG180 | | Descriptor: | (6S,9aS)-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-12-29 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7XBZ
 
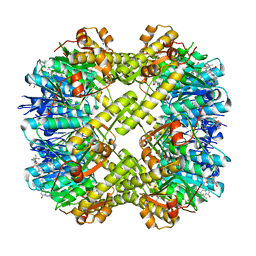 | | Crystal structure of Staphylococcus aureus ClpP in complex with R-ZG197 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-[(1R)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
2CBY
 
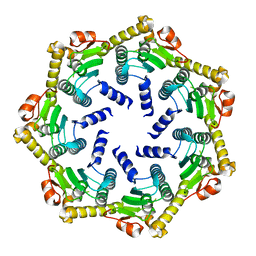 | | Crystal structure of the ATP-dependent Clp Protease proteolytic subunit 1 (ClpP1) from Mycobacterium tuberculosis | | Descriptor: | ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT 1 | | Authors: | Mate, M.J, Portnoi, D, Alzari, P.M, Ortiz-Lombardia, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into the Inter-Ring Plasticity of Caseinolytic Proteases from the X-Ray Structure of Mycobacterium Tuberculosis Clpp1.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2CE3
 
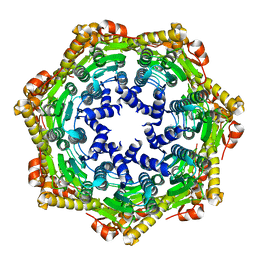 | | CRYSTAL STRUCTURE OF THE ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT 1 (CLPP1) FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT 1 | | Authors: | Segelke, B, Kim, C.Y, Ortiz-Lombardia, M, Alzari, P.M, Lekin, T. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into the Inter-Ring Plasticity of Caseinolytic Proteases from the X-Ray Structure of Mycobacterium Tuberculosis Clpp1.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2FZS
 
 | |
7X8X
 
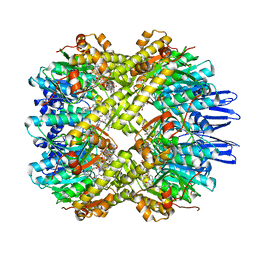 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
5E0S
 
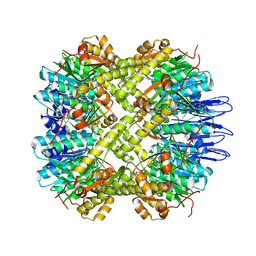 | | crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5DKP
 
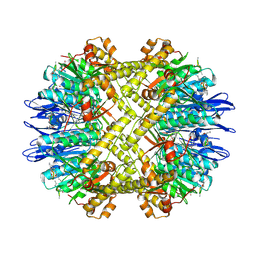 | | Crystal Structure of N. meningitidis ClpP in complex with agonist ADEP A54556. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION, SODIUM ION, ... | | Authors: | Goodreid, J.D, Janetzko, J, Santa Maria Jr, J.P, Wong, K, Leung, E, Eger, B.T, Bryson, S, Pai, E.F, Gray-Owen, S.D, Walker, S, Houry, W.A, Batey, R.A. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Development and Characterization of Potent Cyclic Acyldepsipeptide Analogues with Increased Antimicrobial Activity.
J.Med.Chem., 59, 2016
|
|
2F6I
 
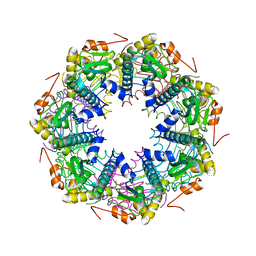 | | Crystal structure of the ClpP protease catalytic domain from Plasmodium falciparum | | Descriptor: | ATP-dependent CLP protease, putative | | Authors: | Mulichak, A, Loppnau, P, Bray, J, Amani, M, Vedadi, M, Wasney, G, Finerty, P, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum.
J.Mol.Biol., 404, 2010
|
|
5DL1
 
 | | ClpP from Staphylococcus aureus in complex with AV145 | | Descriptor: | 1-(propan-2-yl)-N-{[2-(thiophen-2-yl)-1,3-oxazol-4-yl]methyl}-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Vielberg, M.-T, Groll, M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible Inhibitors Arrest ClpP in a Defined Conformational State that Can Be Revoked by ClpX Association.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5C90
 
 | | Staphylococcus aureus ClpP mutant - Y63A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Liu, H, Zhang, J, Gan, J, Yang, C.-G. | | Deposit date: | 2015-06-26 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of Gain-of-Function Mutant Provides New Insights into ClpP Structure
Acs Chem.Biol., 11, 2016
|
|
5DZK
 
 | | Crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, BEZ-LEU-LEU | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
3KTK
 
 | |
3KTH
 
 | |
