5LQ2
 
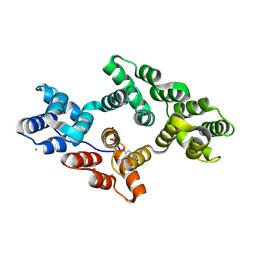 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 3.4 A resolution | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
7WU7
 
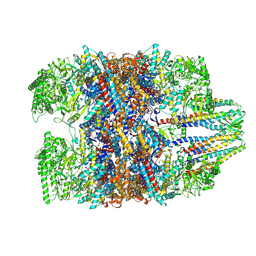 | | Prefoldin-tubulin-TRiC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Prefoldin subunit 1, Prefoldin subunit 2, ... | | Authors: | Gestaut, D, Zhao, Y, Park, J, Ma, B, Leitner, A, Collier, M, Pintilie, G, Roh, S.-H, Chiu, W, Frydman, J. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
2F8X
 
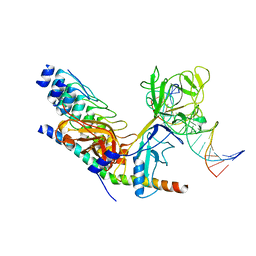 | | Crystal structure of activated Notch, CSL and MAML on HES-1 promoter DNA sequence | | Descriptor: | 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
8JHO
 
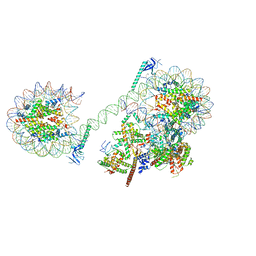 | |
5WVI
 
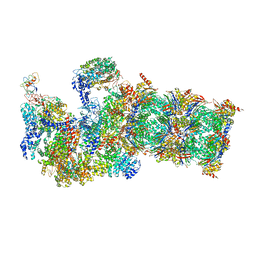 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5LN3
 
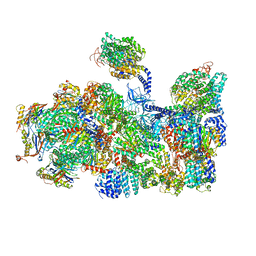 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
4MH8
 
 | |
1W82
 
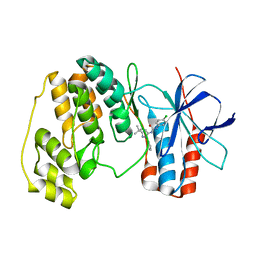 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[(3Z)-5-TERT-BUTYL-2-PHENYL-1,2-DIHYDRO-3H-PYRAZOL-3-YLIDENE]-N'-(4-CHLOROPHENYL)UREA | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation
J.Med.Chem., 48, 2005
|
|
6KW3
 
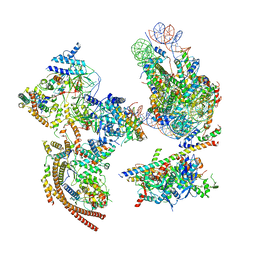 | | The ClassA RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
1W84
 
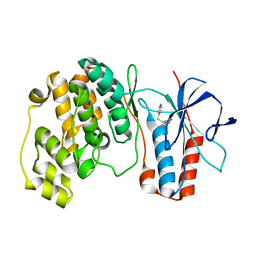 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(2-PYRIDIN-4-YLETHYL)-1H-INDOLE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation.
J. Med. Chem., 48, 2005
|
|
6KW5
 
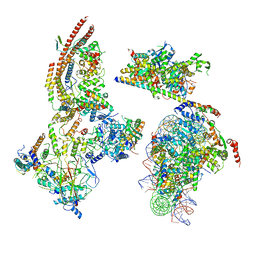 | | The ClassC RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome
To Be Published
|
|
3BV2
 
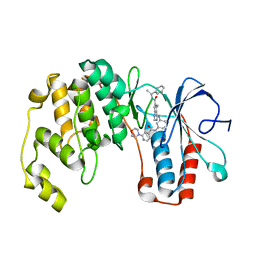 | | Morpholino pyrrolotriazine P38 Alpha map kinase inhibitor compound 30 | | Descriptor: | 5-methyl-4-[(2-methyl-5-{[(2-morpholin-4-ylpyridin-4-yl)carbonyl]amino}phenyl)amino]-N-(1-phenylethenyl)pyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and SAR of new pyrrolo[2,1-f][1,2,4]triazines as potent p38 alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BV3
 
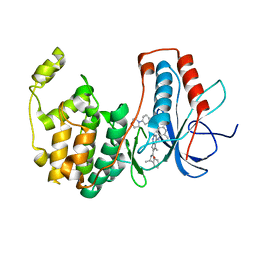 | | Morpholino pyrrolotriazine P38 Alpha Map Kinase inhibitor compound 2 | | Descriptor: | 5-methyl-4-[(2-methyl-5-{[(3-morpholin-4-ylphenyl)carbonyl]amino}phenyl)amino]-N-[(1S)-1-phenylethyl]pyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Synthesis and SAR of new pyrrolo[2,1-f][1,2,4]triazines as potent p38 alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2R29
 
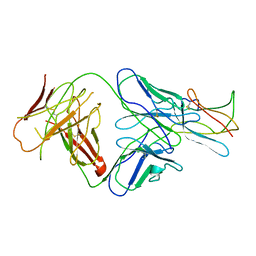 | | Neutralization of dengue virus by a serotype cross-reactive antibody elucidated by cryoelectron microscopy and x-ray crystallography | | Descriptor: | Envelope protein E, Heavy chain of Fab 1A1D-2, Light chain of Fab 1A1D-2 | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-08-24 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5LHG
 
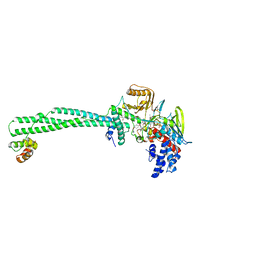 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-methyl-N-[4-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-methyl-N-[4-[[4-[(1-methyl-4-piperidyl)oxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LHI
 
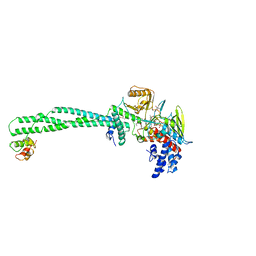 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
5LHH
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-ethyl-N-[3-(methoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-ethyl-~{N}-[3-(methoxymethyl)-2-[[4-[[(3~{R})-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
3KQ7
 
 | |
5WJJ
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, N-{4-[2-(4-fluoro-3-methylphenyl)imidazo[1,2-b]pyridazin-3-yl]pyridin-2-yl}-2-methyl-1-oxo-1lambda~5~-pyridine-4-carboxamide | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-07-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
5LUQ
 
 | | Crystal Structure of Human DNA-dependent Protein Kinase Catalytic Subunit (DNA-PKcs) | | Descriptor: | C-terminal fragment of KU80 (KU80ct194), DNA-dependent protein kinase catalytic subunit,DNA-dependent Protein Kinase Catalytic Subunit,DNA-dependent protein kinase catalytic subunit | | Authors: | Sibanda, B.L, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-15 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair.
Science, 355, 2017
|
|
8TOF
 
 | | Rpd3S bound to an H3K36Cme3 modified nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (176-MER), Histone H2A, ... | | Authors: | Markert, J.W, Vos, S.M, Farnung, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex.
Nat Commun, 14, 2023
|
|
5WKN
 
 | | Crystal structure of the parainfluenza virus 5 nucleoprotein-phosphoprotein complex | | Descriptor: | Nucleoprotein, Phosphoprotein | | Authors: | Aggarwal, M, Leser, G.P, Kors, C.A, Lamb, R.A. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the Paramyxovirus Parainfluenza Virus 5 Nucleoprotein in Complex with an Amino-Terminal Peptide of the Phosphoprotein.
J. Virol., 92, 2018
|
|
1M3Q
 
 | |
1M7Q
 
 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4WZS
 
 | |
