3G3Y
 
 | | Mth0212 in complex with ssDNA in space group P32 | | Descriptor: | 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-03 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3GA6
 
 | | Mth0212 in complex with two DNA helices | | Descriptor: | 5'-D(*GP*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
1ORP
 
 | |
1P59
 
 | | Structure of a non-covalent Endonuclease III-DNA Complex | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*GP*(5IU)P*GP*GP*AP*C)-3', 5'-D(TP*GP*(5IU)P*CP*CP*AP*(3DR)P*GP*(5IU)P*CP*T)-3', Endonuclease III, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-04-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Trapped Endonuclease III-DNA Covalent Intermediate
Embo J., 22, 2003
|
|
7DVD
 
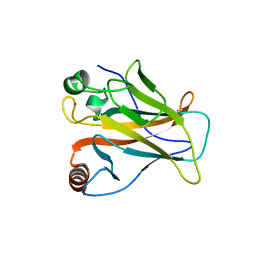 | | The crystal structure of p53 DNA binding domain and PUMA complex | | Descriptor: | Bcl-2-binding component 3, isoforms 1/2, Cellular tumor antigen p53, ... | | Authors: | Han, C.W, Lee, H.N, Jeong, M.S, Jang, S.B. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of the p53 DNA binding domain and PUMA complex.
Biochem.Biophys.Res.Commun., 548, 2021
|
|
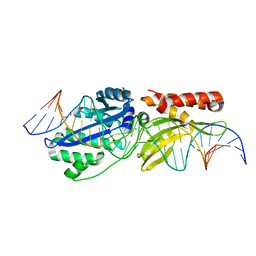
6UVW
 
 | |
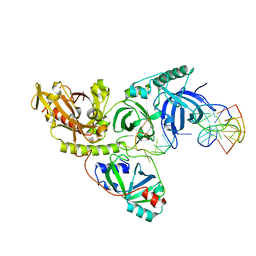
1PHJ
 
 | |
6UWK
 
 | |
1IRZ
 
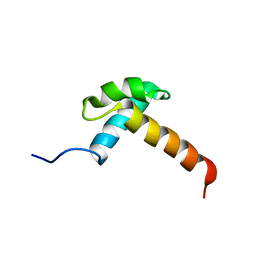 | |
6UWJ
 
 | |
6UWH
 
 | |
4QCL
 
 | | Crystal structure of the catalytic core of human DNA polymerase alpha in ternary complex with an RNA-primed DNA template and dCTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA TEMPLATE, ... | | Authors: | Baranovskiy, A.G, Suwa, Y, Babayeva, N.D, Gu, J, Tahirov, T.H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity and fidelity of human DNA polymerase alpha depend on primer structure
J.Biol.Chem., 2018
|
|
8FDQ
 
 | |
6UWG
 
 | |
6UW0
 
 | |
8INB
 
 | | Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex | | Descriptor: | Cas12j-SF05, NTS-DNA, TS-DNA, ... | | Authors: | Zhang, X, Duan, Z.Q, Zhu, J.K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for DNA cleavage by the hypercompact Cas12j-SF05.
Cell Discov, 9, 2023
|
|
1DSM
 
 | | (-)-duocarmycin SA covalently linked to duplex DNA | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, 5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3' | | Authors: | Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structural basis for in situ activation of DNA alkylation by duocarmycin SA
J.Mol.Biol., 300, 2000
|
|
5J70
 
 | | The Chd1 DNA-binding domain in complex with 17mer DNA duplex | | Descriptor: | Chromo domain-containing protein 1, DNA (5'-D(*CP*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*CP*G)-3') | | Authors: | Bowman, G.D, Jenkins, K.R, Hauk, G. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Chd1 domains communicate across the DNA gyres of the nucleosome
To Be Published
|
|
3PR5
 
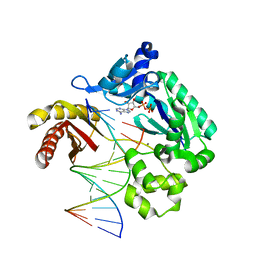 | | Dpo4 Y12A mutant incorporating ADP opposite template dT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Kirouac, K.N, Ling, H. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of ribonucleotide discrimination by a Y-family DNA polymerase.
J.Mol.Biol., 407, 2011
|
|
7LW7
 
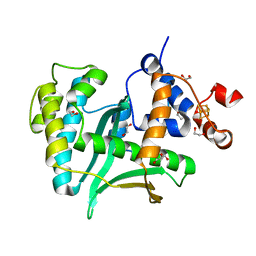 | | Human Exonuclease 5 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Exonuclease V, GLYCEROL, ... | | Authors: | Tsai, C.L, Tainer, J.A. | | Deposit date: | 2021-02-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EXO5-DNA structure and BLM interactions direct DNA resection critical for ATR-dependent replication restart.
Mol.Cell, 81, 2021
|
|
8WH5
 
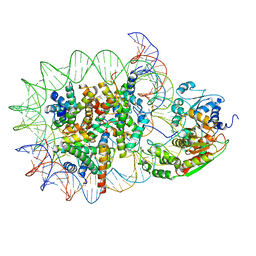 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHA
 
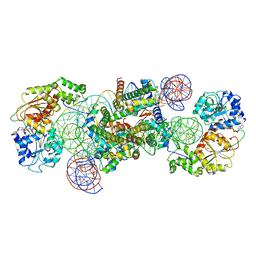 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
4R49
 
 | | Racemic crystal structure of a calcium-bound B-DNA duplex | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8WH8
 
 | | Structure of DDM1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
