1EE9
 
 | | CRYSTAL STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH NAD | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-31 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
1EHF
 
 | |
1ELS
 
 | | CATALYTIC METAL ION BINDING IN ENOLASE: THE CRYSTAL STRUCTURE OF ENOLASE-MN2+-PHOSPHONOACETOHYDROXAMATE COMPLEX AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, MANGANESE (II) ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Zhang, E, Hatada, M, Brewer, J.M, Lebioda, L. | | Deposit date: | 1994-04-05 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution.
Biochemistry, 33, 1994
|
|
2VJB
 
 | | Torpedo Californica Acetylcholinesterase In Complex With A Non Hydrolysable Substrate Analogue, 4-Oxo-N,N,N- Trimethylpentanaminium - Orthorhombic space group - Dataset D at 100K | | Descriptor: | (4R)-4-HYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Bourgeois, D, Fournier, D, Silman, I, Sussman, J.L, Weik, M. | | Deposit date: | 2007-12-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6N4R
 
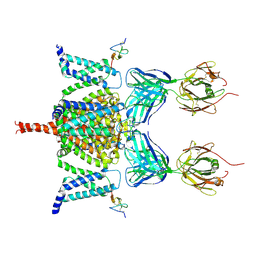 | | CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
1EDZ
 
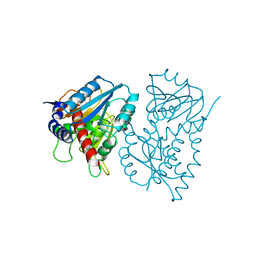 | | STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-28 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
7O5R
 
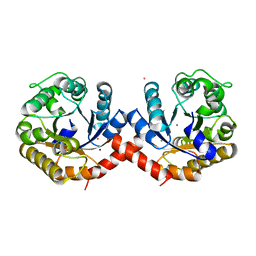 | | Crystal structure of holo-SwHPA-Mn (hydroxyketoacid aldolase) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7P1Z
 
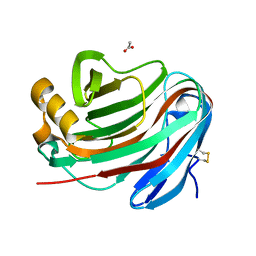 | | Novel GH12 endogluconase from Aspergillus cervinus | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase, ... | | Authors: | Lazarenko, V.A, Rykov, S.V, Nikolaeva, A.Y, Berezina, O.V, Akentyev, F.I. | | Deposit date: | 2021-07-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Unusual substrate specificity in GH family 12: structure-function analysis of glucanases Bgh12A and Xgh12B from Aspergillus cervinus, and Egh12 from Thielavia terrestris.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7T2F
 
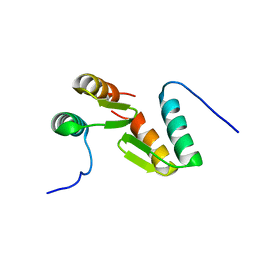 | | Solution structure of the model HEEH mini protein homodimer HEEH_TK_rd5_0341 | | Descriptor: | HEEH mini protein HEEH_TK_rd5_0341 | | Authors: | Lemak, A, Houliston, S, Kim, T.-E, Martel, C, Rocklin, G.J, Arrowsmith, C.H. | | Deposit date: | 2021-12-04 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dissecting the stability determinants of a challenging de novo protein fold using massively parallel design and experimentation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SR6
 
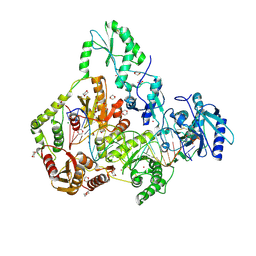 | | Human Endogenous Retrovirus (HERV-K) reverse transcriptase ternary complex with dsDNA template Primer and dNTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Baldwin, E.T, Nichols, C. | | Deposit date: | 2021-11-08 | | Release date: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K-specific inhibition.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TB1
 
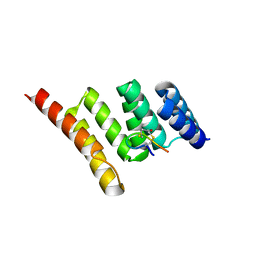 | | Crystal structure of STUB1 with a macrocyclic peptide | | Descriptor: | 1,3-bis(sulfanyl)propan-2-one, ALA-CYS-SER-SER-ILE-TRP-CYS-PRO-ASP-GLY, E3 ubiquitin-protein ligase CHIP | | Authors: | Bahmanjah, S, Klein, D.J. | | Deposit date: | 2021-12-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Discovery and Structure-Based Design of Macrocyclic Peptides Targeting STUB1.
J.Med.Chem., 2022
|
|
7SLT
 
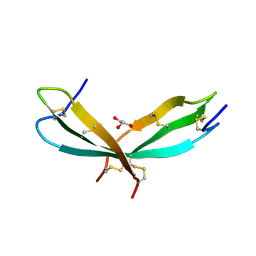 | |
7SND
 
 | | Pacifastin related protease inhibitors | | Descriptor: | GLYCEROL, PHOSPHATE ION, Pacifastin-related peptide | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
6NNV
 
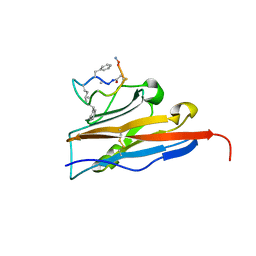 | | PD-L1 IgV domain complex with macro-cyclic peptide | | Descriptor: | Programmed cell death 1 ligand 1, macrocyclic peptide | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-15 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7UG6
 
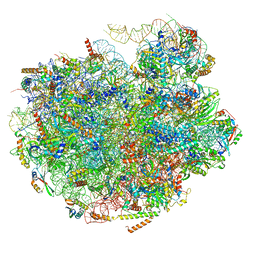 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2VT6
 
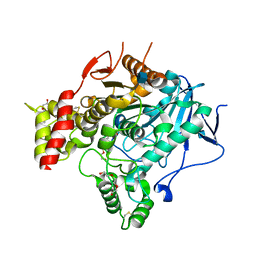 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 9400000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4JDJ
 
 | |
4JRY
 
 | | Crystal Structure of SB47 TCR-HLA B*3505-LPEP complex | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Highly divergent T-cell receptor binding modes underlie specific recognition of a bulged viral peptide bound to a human leukocyte antigen class I molecule.
J.Biol.Chem., 288, 2013
|
|
7Q2X
 
 | | Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (Form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Condensin complex subunit 1, ... | | Authors: | Lee, B.-G, Rhodes, J, Lowe, J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Clamping of DNA shuts the condensin neck gate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PC4
 
 | | The PDZ domain of SNTB1 complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-1-syntrophin,Annexin A2, CALCIUM ION, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC5
 
 | | The third PDZ domain of PDZD7 complexed with the PDZ-binding motif of EXOC4 | | Descriptor: | CALCIUM ION, Exocyst complex component 4, GLYCEROL, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PCB
 
 | | The PDZ domain of SNX27 fused with ANXA2 | | Descriptor: | CALCIUM ION, GLYCEROL, Sorting nexin-27,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC7
 
 | | The PDZ domain of SNTG1 complexed with the acetylated PDZ-binding motif of PTEN | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC9
 
 | | The PDZ domain of SYNJ2BP complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, Protein Tax-1, Synaptojanin-2-binding protein,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC3
 
 | | The second PDZ domain of DLG1 complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, Disks large homolog 1,Annexin A2, GLYCEROL, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
