7M63
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-70099 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[(1R,3S,5S,6r)-3-(5,6-difluoro-1H-benzimidazol-1-yl)bicyclo[3.1.0]hexan-6-yl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
6DPR
 
 | | Mapping the binding trajectory of a suicide inhibitor in human indoleamine 2,3-dioxygenase 1 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[cis-4-(6-fluoroquinolin-4-yl)cyclohexyl]propanamide, 1,2-ETHANEDIOL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Pham, K.N, Yeh, S.R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mapping the Binding Trajectory of a Suicide Inhibitor in Human Indoleamine 2,3-Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
6DPQ
 
 | | Mapping the binding trajectory of a suicide inhibitor in human indoleamine 2,3-dioxygenase 1 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[cis-4-(6-fluoroquinolin-4-yl)cyclohexyl]propanamide, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Pham, K.N, Yeh, S.R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Mapping the Binding Trajectory of a Suicide Inhibitor in Human Indoleamine 2,3-Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
7M7D
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-8968 | | Descriptor: | (5S)-6,6-dimethyl-8-[(4S)-7-(trifluoromethyl)imidazo[1,5-a]pyridin-5-yl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
6E40
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complexed with ferric heme and Epacadostat | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E41
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and an Epacadostat analog | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]sulfanyl}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E43
 
 | | Crystal structure of human indoleamine 2,3-dioxygenase 1 (IDO1) in complex with a BMS-978587 analog | | Descriptor: | (1R,2S)-2-(4-[cyclohexyl(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, BENZOIC ACID, Indoleamine 2,3-dioxygenase 1 | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E45
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferrous state | | Descriptor: | GLYCEROL, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3TH1
 
 | | Crystal structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ACETATE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Rustiguel, J.K, Nonato, M.C. | | Deposit date: | 2011-08-18 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida
To be Published
|
|
5FZ7
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment ethyl 2-amino-4-thiophen-2-ylthiophene-3- carboxylate (N06131b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Ethyl 2-Amino-4-Thiophen-2-Ylthiophene-3-Carboxylate (N06131B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5M8R
 
 | | Crystal structure of human tyrosinase related protein 1 (T391V-R374S-Y362F) in complex with mimosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, X, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2016-10-29 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human Tyrosinase Related Protein 1 Reveals a Binuclear Zinc Active Site Important for Melanogenesis.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3JSX
 
 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | Descriptor: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Dunstan, M.S, Levy, C, Leys, D. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
1LUC
 
 | | BACTERIAL LUCIFERASE | | Descriptor: | 1,2-ETHANEDIOL, BACTERIAL LUCIFERASE, MAGNESIUM ION | | Authors: | Fisher, A.J, Rayment, I. | | Deposit date: | 1996-05-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A resolution crystal structure of bacterial luciferase in low salt conditions.
J.Biol.Chem., 271, 1996
|
|
6CR2
 
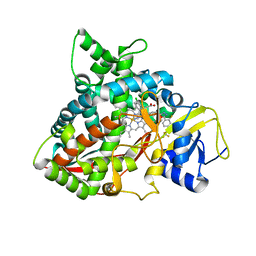 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from Aspergillus fumigatus in complex with the VNI derivative N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | 14-alpha sterol demethylase Cyp51B, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2018-03-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Sterol 14 alpha-Demethylase Structure-Based Design of VNI (( R)- N-(1-(2,4-Dichlorophenyl)-2-(1 H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide)) Derivatives To Target Fungal Infections: Synthesis, Biological Evaluation, and Crystallographic Analysis.
J. Med. Chem., 61, 2018
|
|
5M8N
 
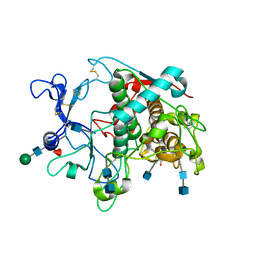 | | Crystal structure of human tyrosinase related protein 1 in complex with mimosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, X, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2016-10-29 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Tyrosinase Related Protein 1 Reveals a Binuclear Zinc Active Site Important for Melanogenesis.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5FZK
 
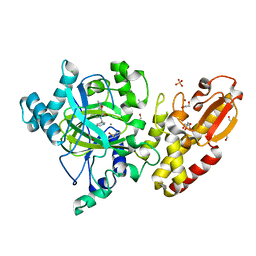 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment N,3-dimethyl-N-(pyridin-3-ylmethyl)-1,2-oxazole-5- carboxamide (N10051a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Talon, R, Collins, P, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with 3D Fragment N,3-Dimethyl-N-(Pyridin-3-Ylmethyl)-1,2-Oxazole-5-Carboxamide (N10051A) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5M8O
 
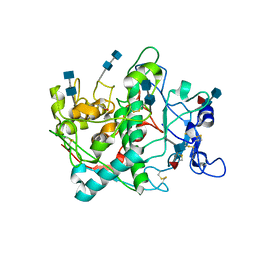 | | Crystal structure of human tyrosinase related protein 1 in complex with tropolone | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, X, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2016-10-29 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Tyrosinase Related Protein 1 Reveals a Binuclear Zinc Active Site Important for Melanogenesis.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5M8T
 
 | | Crystal structure of human tyrosinase related protein 1 (T391V-R374S-Y362F) in complex with tropolone | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, X, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2016-10-29 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Human Tyrosinase Related Protein 1 Reveals a Binuclear Zinc Active Site Important for Melanogenesis.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4A27
 
 | | Crystal structure of human synaptic vesicle membrane protein VAT-1 homolog-like protein | | Descriptor: | 1,2-ETHANEDIOL, SYNAPTIC VESICLE MEMBRANE PROTEIN VAT-1 HOMOLOG-LIKE | | Authors: | Vollmar, M, Shafqat, N, Muniz, J.R.C, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Synaptic Vesicle Membrane Protein Vat-1 Homolog-Like Protein
To be Published
|
|
5FZL
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 3-methyl-N-pyridin-4-yl-1,2-oxazole-5-carboxamide (N09954a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(pyridin-4-yl)-1,2-oxazole-5-carboxamide, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with 3D Fragment 3-Methyl-N-Pyridin-4-Yl-1,2-Oxazole-5-Carboxamide (N09954A) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5FZ6
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment N05859b (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-CARBOXYPIPERIDINE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment N05859B (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5FZM
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 5-(2-fluorophenyl)-1,3-oxazole-4-carboxylic acid (N09989b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-fluorophenyl)-1,3-oxazole-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with 3D Fragment 5-(2-Fluorophenyl)-1,3-Oxazole-4-Carboxylic Acid (N09989B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
3N9T
 
 | | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4 | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, CITRATE ANION, FE (III) ION, ... | | Authors: | Liu, W, Shen, W, Fang, P, Li, J, Cui, Z. | | Deposit date: | 2010-05-31 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4
To be Published
|
|
5FZB
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 4-Pyridylthiourea (N06275b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-pyridin-4-ylthiourea, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment 4-Pyridylthiourea (N06275B) (Ligand Modelled Based on Pandda Event Map, Sgc -Diamond I04-1 Fragment Screening)
To be Published
|
|
5FZ9
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment thieno(3,2-b)thiophene-5-carboxylic acid (N06263b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Thieno(3,2-B)Thiophene -5-Carboxylic Acid (N06263B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
