7O3Q
 
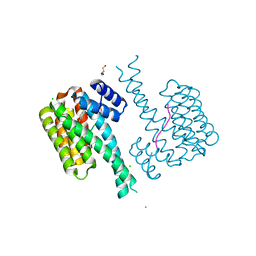 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-041 | | Descriptor: | 14-3-3 protein sigma, 4-methyl-~{N}-(1-methylpyrazol-3-yl)benzenesulfonamide, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O6I
 
 | |
7O5A
 
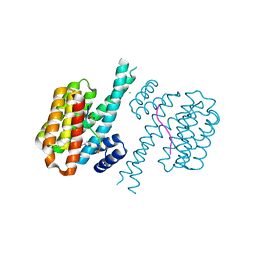 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-158 | | Descriptor: | 14-3-3 protein sigma, 4-(3-methoxyazetidin-1-yl)carbonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O6O
 
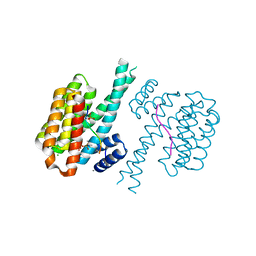 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-096 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) propane-2-sulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O6G
 
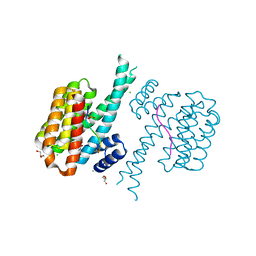 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-176 | | Descriptor: | 14-3-3 protein sigma, 4-[(3~{R})-3-oxidanylpiperidin-1-yl]carbonylbenzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NSV
 
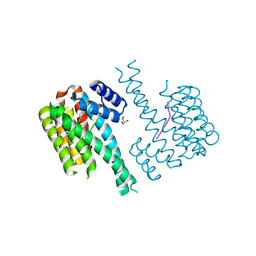 | | 14-3-3 sigma with p65 (RelA) binding site pS45 and covalently bound PC2046 | | Descriptor: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, GLYCEROL, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
8A4L
 
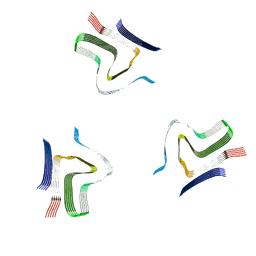 | | Lipidic alpha-synuclein fibril - polymorph L2A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-06-12 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
1RUD
 
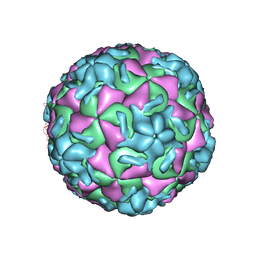 | | RHINOVIRUS 14 MUTANT N1105S COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52084 | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUE
 
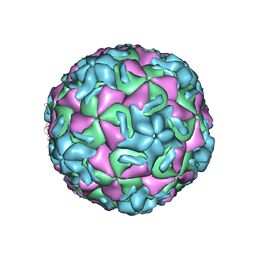 | | RHINOVIRUS 14 SITE DIRECTED MUTANT N1219A COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52035 | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUH
 
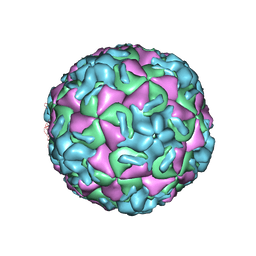 | | RHINOVIRUS 14 MUTANT N1219S COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52084 | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
2HN7
 
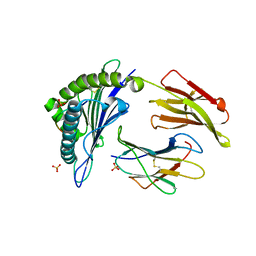 | | HLA-A*1101 in complex with HBV peptide homologue | | Descriptor: | Beta-2-microglobulin, DNA polymerase PEPTIDE HOMOLOGUE, HLA class I histocompatibility antigen, ... | | Authors: | Blicher, T. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of HLA-A*1101 in complex with a hepatitis B peptide homologue.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5YK5
 
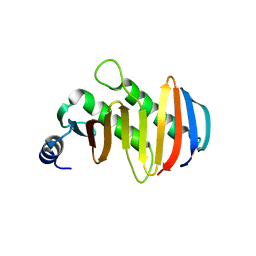 | | structure of the human Lamtor4-Lamtor5 complex | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Wu, G, Mu, Z. | | Deposit date: | 2017-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane
Cell Discov, 3, 2017
|
|
7QJ9
 
 | |
7QJ6
 
 | |
1BEV
 
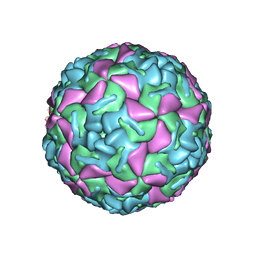 | | BOVINE ENTEROVIRUS VG-5-27 | | Descriptor: | BOVINE ENTEROVIRUS COAT PROTEINS VP1 TO VP4, MYRISTIC ACID, SULFATE ION | | Authors: | Smyth, M, Tate, J, Lyons, C, Hoey, E, Martin, S, Stuart, D. | | Deposit date: | 1996-04-03 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Implications for viral uncoating from the structure of bovine enterovirus.
Nat.Struct.Biol., 2, 1995
|
|
1CC0
 
 | | CRYSTAL STRUCTURE OF THE RHOA.GDP-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, rho GDP dissociation inhibitor alpha, ... | | Authors: | Longenecker, K.L, Read, P, Derewenda, U, Dauter, Z, Garrard, S, Walker, L, Somlyo, A.V, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 1999-03-03 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | How RhoGDI binds Rho.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3O7L
 
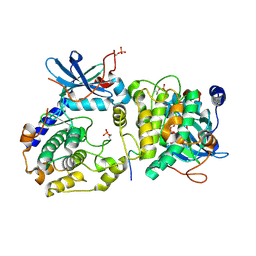 | |
5SYT
 
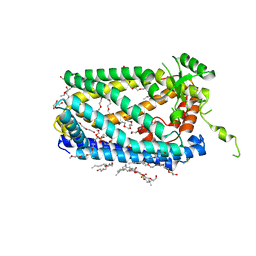 | | Crystal Structure of ZMPSTE24 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX prenyl protease 1 homolog, ... | | Authors: | Clark, K, Jenkins, J.L, Fedoriw, N, Dumont, M.E, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors.
Protein Sci., 26, 2017
|
|
5ZMC
 
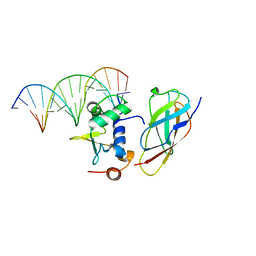 | | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*GP*AP*CP*CP*CP*GP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*GP*CP*CP*CP*TP*TP*CP*CP*GP*GP*GP*TP*CP*CP*CP*C)-3'), Nuclear factor NF-kappa-B p100 subunit, ... | | Authors: | Xu, X, Bharath, S.R, Song, H. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Nat Commun, 9, 2018
|
|
2PK2
 
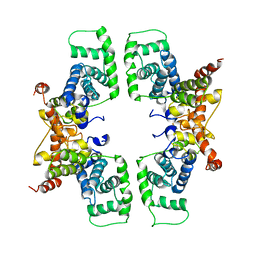 | | Cyclin box structure of the P-TEFb subunit Cyclin T1 derived from a fusion complex with EIAV Tat | | Descriptor: | Cyclin-T1, Protein Tat | | Authors: | Anand, K, Schulte, A, Fujinaga, K, Scheffzek, K, Geyer, M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Cyclin Box Structure of the P-TEFb Subunit Cyclin T1 Derived from a Fusion Complex with EIAV Tat.
J.Mol.Biol., 370, 2007
|
|
1SYK
 
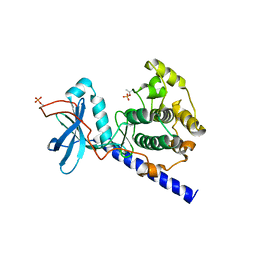 | | Crystal structure of E230Q mutant of cAMP-dependent protein kinase reveals unexpected apoenzyme conformation | | Descriptor: | cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Wu, J, Yang, J, Madhusudan, N, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the E230Q mutant of cAMP-dependent
protein kinase reveals an unexpected apoenzyme conformation and an
extended N-terminal A helix.
Protein Sci., 14, 2005
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
1YRN
 
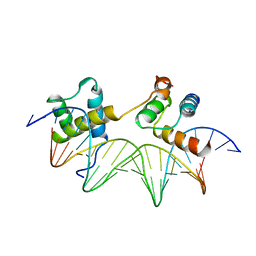 | | CRYSTAL STRUCTURE OF THE MATA1/MATALPHA2 HOMEODOMAIN HETERODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*TP*AP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*TP*AP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT A1 HOMEODOMAIN), ... | | Authors: | Li, T, Stark, M.R, Johnson, A.D, Wolberger, C. | | Deposit date: | 1995-11-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA.
Science, 270, 1995
|
|
7C9O
 
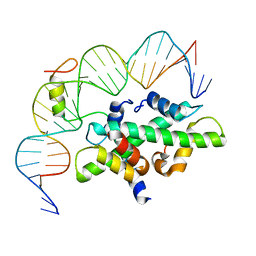 | | Crystal structure of DNA-bound CCT/NF-YB/YC complex (HD1CCT/GHD8/OsNF-YC2) | | Descriptor: | DNA (25-MER), Nuclear transcription factor Y subunit B-11, Nuclear transcription factor Y subunit C-2, ... | | Authors: | Shen, C, Liu, H, Guan, Z, Xing, Y, Yin, P. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering.
Plant Cell, 32, 2020
|
|
1Q71
 
 | | The structure of microcin J25 is a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Clark, R, Daly, N.L, Goransson, U, Jones, A, Craik, D.J. | | Deposit date: | 2003-08-14 | | Release date: | 2003-12-16 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone.
J.Am.Chem.Soc., 125, 2003
|
|
