3AB6
 
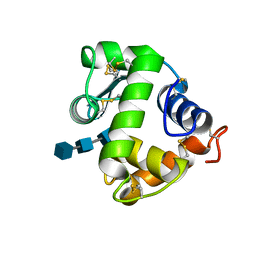 | | Crystal structure of NAG3 bound lysozyme from Meretrix lusoria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme | | Authors: | Yoneda, K, Kuwano, Y, Araki, T. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The tertiary structure of an i-type lysozyme isolated from the common orient clam (Meretrix lusoria)
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ZW8
 
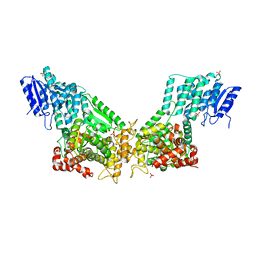 | | Crystal Structure Of Rat Peroxisomal Multifunctional Enzyme Type 1 (rpMFE1) In Apo Form | | Descriptor: | GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, SULFATE ION | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZWB
 
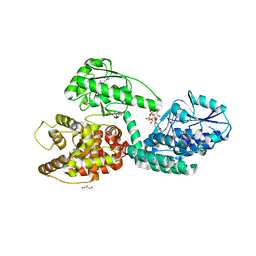 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 2TRANS-HEXENOYL-COA | | Descriptor: | (2E)-Hexenoyl-CoA, GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZWC
 
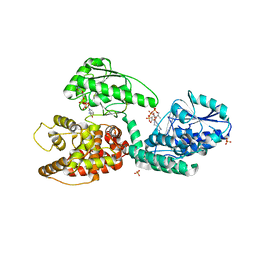 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-DECANOYL-COA | | Descriptor: | (S)-3-HYDROXYDECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3VZK
 
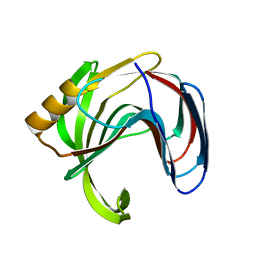 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZO
 
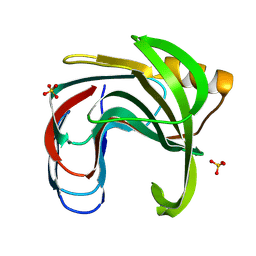 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
1ZYT
 
 | | Crystal structure of spin labeled T4 Lysozyme (A82R1) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | EPR (1.7 Å), X-RAY DIFFRACTION | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins
Protein Sci., 18, 2009
|
|
3ZW9
 
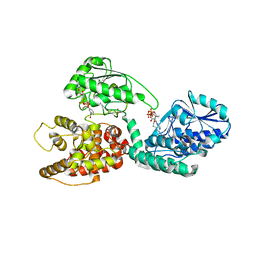 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA | | Descriptor: | (2S,3S)-3-HYDROXY-2-METHYLBUTANOYL-COA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
2E6D
 
 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with fumarate | | Descriptor: | COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Inaoka, D.K, Shimizu, H, Sakamoto, K, Shiba, T, Kurisu, G, Nara, T, Aoki, T, Harada, S, Kita, K. | | Deposit date: | 2006-12-26 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of Trypanosoma cruzi dihydroorotate dehydrogenase complexed with substrates and products: atomic resolution insights into mechanisms of dihydroorotate oxidation and fumarate reduction
Biochemistry, 47, 2008
|
|
3KJ0
 
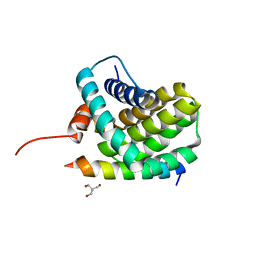 | | Mcl-1 in complex with Bim BH3 mutant I2dY | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KJ1
 
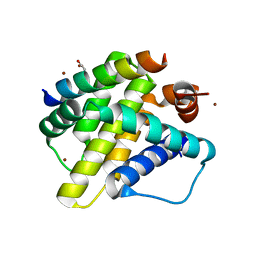 | | Mcl-1 in complex with Bim BH3 mutant I2dA | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, CHLORIDE ION, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
6JLV
 
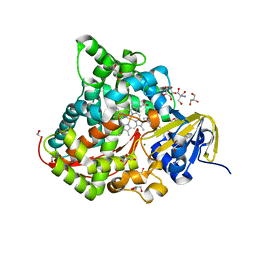 | | Near-Atomic Resolution Structure of the CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JMW
 
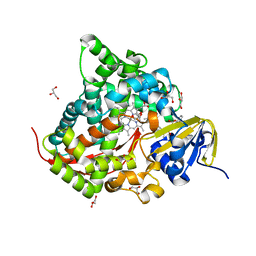 | | Structure of the Chromium Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, Chromium Protoporphyrin IX, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JMH
 
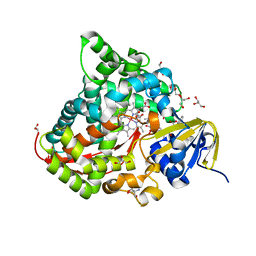 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JO1
 
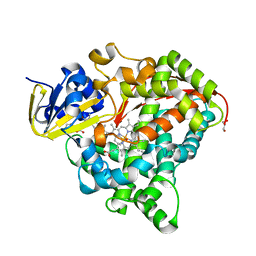 | | Structure of the CYP102A1 Haem Domain with N-(S)-Ibuprofenoyl-L-Phenylalanine | | Descriptor: | (2S)-2-[[(2S)-2-[4-(2-methylpropyl)phenyl]propanoyl]amino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-19 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JS8
 
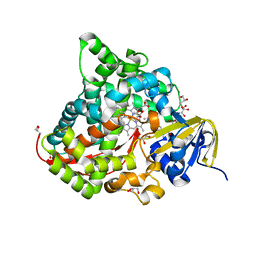 | | Structure of the CYP102A1 Haem Domain with N-Dehydroabietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aS,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,9,10,10a-hexahydrophenanthren-1-yl]carbonylamino]-3-(1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-04-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5U3H
 
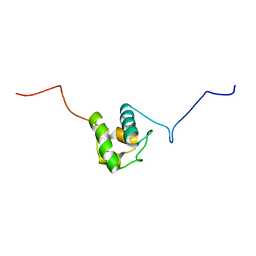 | |
6K58
 
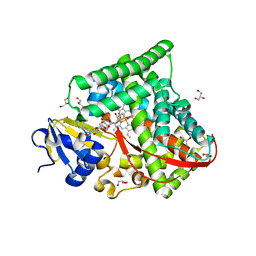 | | Structure of the CYP102A1 Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JVC
 
 | | Structure of the Cobalt Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-04-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3A51
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution with bound 25-hydroxyvitamin D3 | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4H
 
 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (orthorhombic crystal form) | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
6K24
 
 | | Structure of the Rhodium Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1VET
 
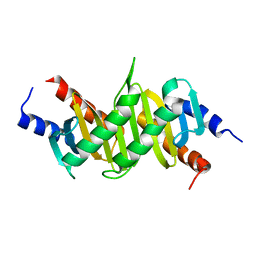 | | Crystal Structure of p14/MP1 at 1.9 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3A4G
 
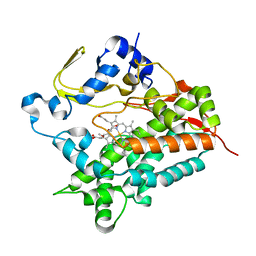 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (trigonal crystal form) | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
1VEU
 
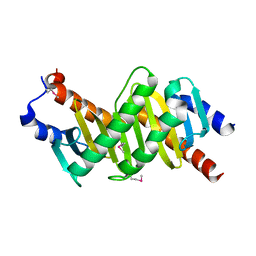 | | Crystal structure of the p14/MP1 complex at 2.15 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
