5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5L33
 
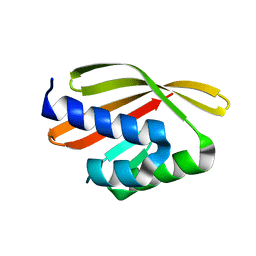 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5KPH
 
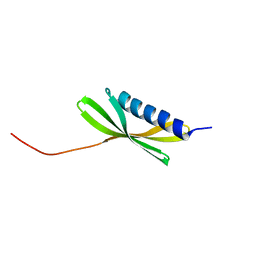 | |
5KPE
 
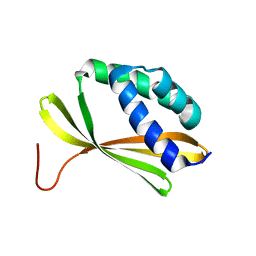 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | Descriptor: | De novo Beta Sheet Design Protein OR664 | | Authors: | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
8QNJ
 
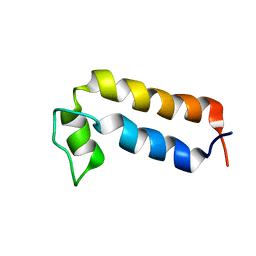 | |
6UY4
 
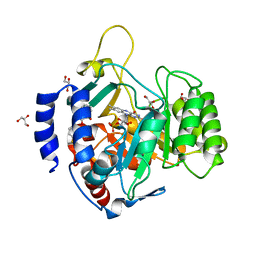 | | Crystal structure of dihydroorotate dehydrogenase from Schistosoma mansoni | | Descriptor: | 2-[(4-fluorophenyl)amino]-3-hydroxynaphthalene-1,4-dione, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mori, R.M, Zapata, L.C.C, Nonato, M.C. | | Deposit date: | 2019-11-11 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural basis for the function and inhibition of dihydroorotate dehydrogenase from Schistosoma mansoni.
Febs J., 288, 2021
|
|
6KOS
 
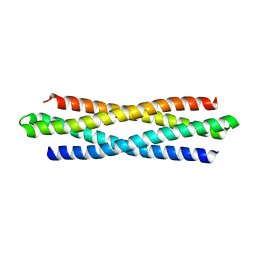 | |
5BOP
 
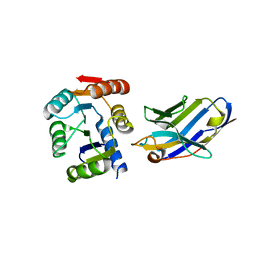 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
1K43
 
 | | 10 Structure Ensemble of the 14-residue peptide RG-KWTY-NG-ITYE-GR (MBH12) | | Descriptor: | MBH12 | | Authors: | Pastor, M.T, Lopez de la Paz, M, Lacroix, E, Serrano, L, Perez-Paya, E. | | Deposit date: | 2001-10-05 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5TRV
 
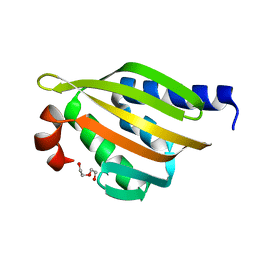 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
9BK5
 
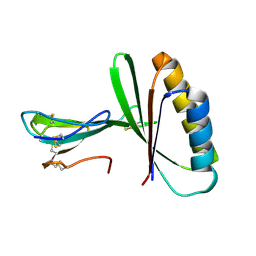 | |
9BK6
 
 | |
6L0L
 
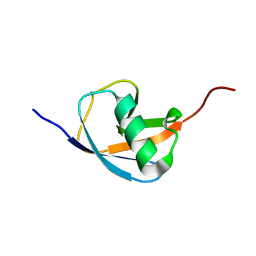 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
9BK7
 
 | |
7VU4
 
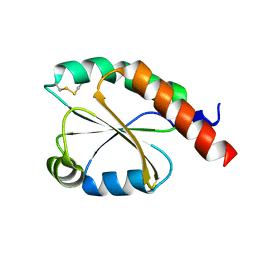 | | de novo design based on 1r26 | | Descriptor: | de novo design protein | | Authors: | Zhang, L. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQW
 
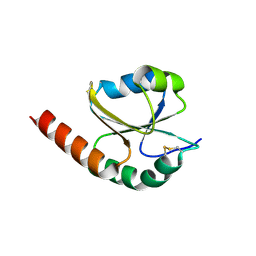 | | de novo designed protein based on 1r26 | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
1K4G
 
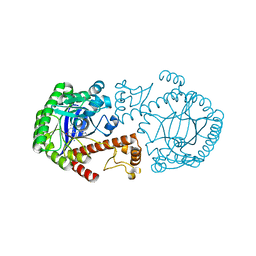 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
1K4H
 
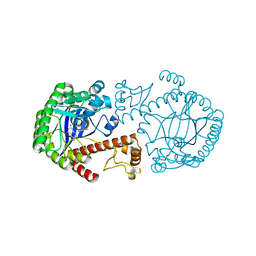 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-Diamino-8-propylsulfanylmethyl-3H-quinazoline-4-one | | Descriptor: | 2,6-DIAMINO-8-PROPYLSULFANYLMETHYL-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE-TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
Chembiochem, 3, 2002
|
|
8XYW
 
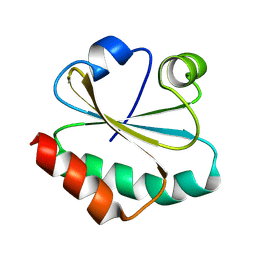 | | De novo designed protein Trx-3 | | Descriptor: | De novo designed Trx-3 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYR
 
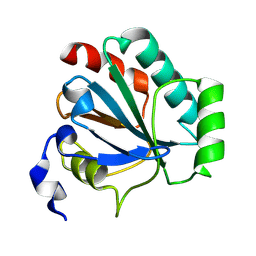 | | De novo designed protein GPX4-2 | | Descriptor: | De novo designed GPX4-2 | | Authors: | Liu, L.J, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYU
 
 | | De novo designed protein GPX4-3 | | Descriptor: | De novo designed GPX4-3 | | Authors: | Guo, Z, Liu, J.L, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYS
 
 | | De novo designed protein GPX4-1 | | Descriptor: | De novo designed GPX4-1 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYT
 
 | | De novo designed protein GPX4-4 | | Descriptor: | De novo designed GPX4-4 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
6L9X
 
 | | Xenons in frog EPDR1 | | Descriptor: | Ependymin-related 1, XENON | | Authors: | Park, S. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | De novo Phasing Xenons Observed in the Frog Ependymin-Related Protein
Crystals, 10, 2020
|
|
