2J4K
 
 | | Crystal structure of uridylate kinase from Sulfolobus solfataricus in complex with UMP to 2.2 Angstrom resolution | | Descriptor: | CADMIUM ION, MAGNESIUM ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Jensen, K.S, Johansson, E, Jensen, K.F. | | Deposit date: | 2006-09-01 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Enzymatic Investigation of the Sulfolobus Solfataricus Uridylate Kinase Shows Competitive Utp Inhibition and the Lack of GTP Stimulation
Biochemistry, 46, 2007
|
|
1S07
 
 | | Crystal Structure of the R253A Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ISOPROPYL ALCOHOL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1UZH
 
 | | A CHIMERIC CHLAMYDOMONAS, SYNECHOCOCCUS RUBISCO ENZYME | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-03-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chimeric Small Subunits Influence Catalysis without Causing Global Conformational Changes in the Crystal Structure of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
1V0H
 
 | | ASCOBATE PEROXIDASE FROM SOYBEAN CYTOSOL IN COMPLEX WITH SALICYLHYDROXAMIC ACID | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SALICYLHYDROXAMIC ACID, ... | | Authors: | Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Ascorbate Peroxidase-Salicylhydroxamic Acid Complex
Biochemistry, 43, 2004
|
|
5L86
 
 | | engineered ascorbate peroxidise | | Descriptor: | Ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hayashi, T, Mittl, P, Hilvert, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Chemically Programmed Proximal Ligand Enhances the Catalytic Properties of a Heme Enzyme.
J. Am. Chem. Soc., 138, 2016
|
|
1XXX
 
 | | Crystal structure of Dihydrodipicolinate Synthase (DapA, Rv2753c) from Mycobacterium tuberculosis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Dihydrodipicolinate synthase, ... | | Authors: | Kefala, G, Panjikar, S, Janowski, R, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-09 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure and kinetic study of dihydrodipicolinate synthase from Mycobacterium tuberculosis.
Biochem.J., 411, 2008
|
|
5DF5
 
 | | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution.
To Be Published
|
|
8PUQ
 
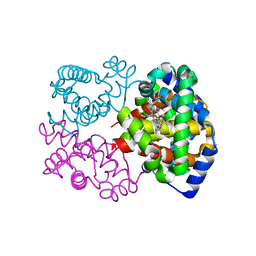 | | MetHemoglobin structure from serial synchrotron crystallography with fixed target | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bjelcic, M, Sigfridsson Clauss, K, Aurelius, O, Milas, M, Nan, J, Ursby, T. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anaerobic fixed-target serial crystallography using sandwiched silicon nitride membranes.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8PUR
 
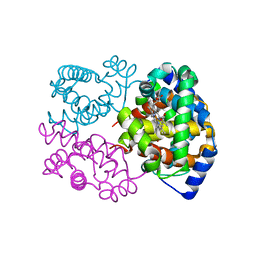 | | DexyHemoglobin structure from serial synchrotron crystallography with fixed target | | Descriptor: | Hemoglobin subunit beta, Hemoglobin, alpha 1, ... | | Authors: | Bjelcic, M, Sigfridsson Clauss, K, Aurelius, O, Milas, M, Nan, J, Ursby, T. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anaerobic fixed-target serial crystallography using sandwiched silicon nitride membranes.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3GBA
 
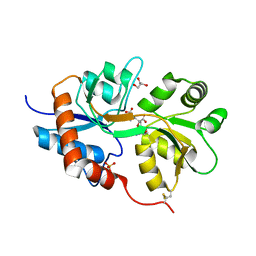 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with dysiherbaine at 1.35A resolution | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
8P1C
 
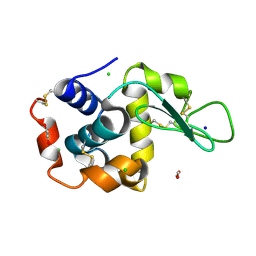 | | Lysozyme structure solved from serial crystallography data collected at 1 kHz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1B
 
 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1A
 
 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz for 5 seconds with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1D
 
 | | Lysozyme structure solved from serial crystallography data collected at 100 Hz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
1IPJ
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS COMPLEXES WITH N-ACETYL-D-GLUCOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
5EEO
 
 | | soybean lipoxygenase(L1)-T756R | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Mikami, B, Nagaya, T, Hioki, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for specificity in lipoxygenase catalysis
To Be Published
|
|
1IPK
 
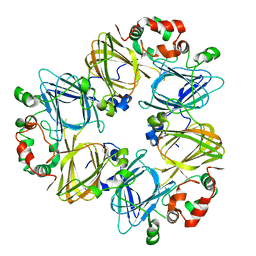 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
3BGD
 
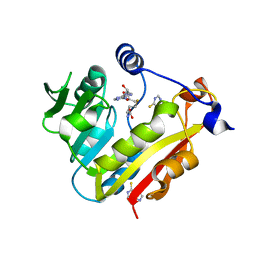 | | Thiopurine S-Methyltransferase | | Descriptor: | 9H-purine-6-thiol, S-ADENOSYL-L-HOMOCYSTEINE, Thiopurine S-methyltransferase | | Authors: | Peng, Y, Yee, V.C. | | Deposit date: | 2007-11-26 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Substrate Recognition in Thiopurine S-Methyltransferase
Biochemistry, 47 (23), 6216 6225, 2008. 10.1021/bi800102x, 2008
|
|
3BGI
 
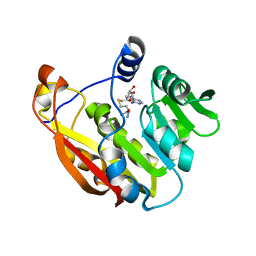 | | Thiopurine S-Methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Thiopurine S-methyltransferase | | Authors: | Peng, Y, Yee, V.C. | | Deposit date: | 2007-11-26 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Substrate Recognition in Thiopurine S-Methyltransferase
Biochemistry, 47 (23), 6216 6225, 2008. 10.1021/bi800102x, 2008
|
|
1HU9
 
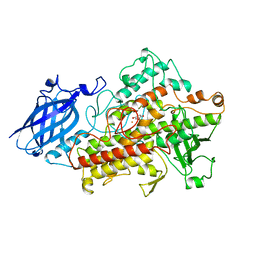 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 4-HYDROPEROXY-2-METHOXY-PHENOL | | Descriptor: | 4-HYDROPEROXY-2-METHOXY-PHENOL, FE (III) ION, LIPOXYGENASE-3 | | Authors: | Zhou, K, Skrzypczak-Jankun, E, McCabe, N.P, Selman, S.H, Jankun, J. | | Deposit date: | 2001-01-04 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of curcumin in complex with lipoxygenase and its significance in
cancer.
INT.J.MOL.MED., 12, 2003
|
|
1JNQ
 
 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH EPIGALLOCATHECHIN (EGC) | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, FE (II) ION, lipoxygenase-3 | | Authors: | Zhou, K, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2001-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of lipoxygenase by (-)-epigallocatechin gallate: X-ray analysis at 2.1 A reveals degradation of EGCG
and shows soybean LOX-3 complex with EGC instead.
INT.J.MOL.MED., 12, 2003
|
|
1IK3
 
 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 13(S)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID | | Descriptor: | (TRANS-12,13-EPOXY)-11-HYDROXY-9(Z)-OCTADECENOIC ACID, (TRANS-12,13-EPOXY)-9-HYDROXY-10(E)-OCTADECENOIC ACID, 13(R)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID, ... | | Authors: | Skrzypczak-Jankun, E, Funk Jr, M.O. | | Deposit date: | 2001-05-01 | | Release date: | 2001-11-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a purple lipoxygenase.
J.Am.Chem.Soc., 123, 2001
|
|
1K9B
 
 | | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28 nm resolution. Structural peculiarities in a folded protein conformation | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Voss, R.H, Ermler, U, Essen, L.O, Wenzl, G, Kim, Y.M, Flecker, P. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28-nm resolution. Structural peculiarities in a folded protein conformation.
Eur.J.Biochem., 242, 1996
|
|
1HYP
 
 | |
1PI2
 
 | |
