4CM3
 
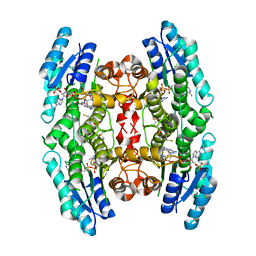 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5-phenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CL8
 
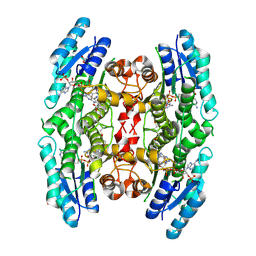 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 2,4-diamino-6-(3-formylphenyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CMI
 
 | |
4CM4
 
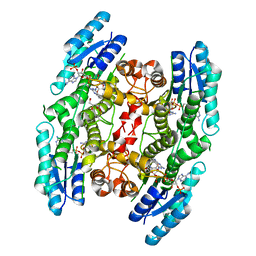 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 5-(4-fluorophenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, GLYCEROL, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4CLO
 
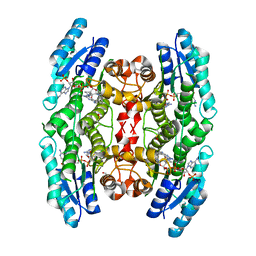 | |
2VKV
 
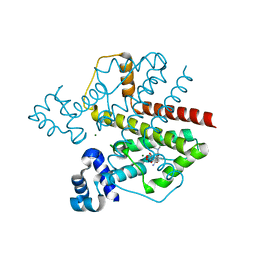 | | TetR (BD) variant L17G with reverse phenotype | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, MAGNESIUM ION, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, ... | | Authors: | Resch, M, Striegl, H, Henssler, E.M, Sevvana, M, Egerer-Sieber, C, Schiltz, E, Hillen, W, Muller, Y.A. | | Deposit date: | 2008-01-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Protein Functional Leap: How a Single Mutation Reverses the Function of the Transcription Regulator Tetr.
Nucleic Acids Res., 36, 2008
|
|
5N68
 
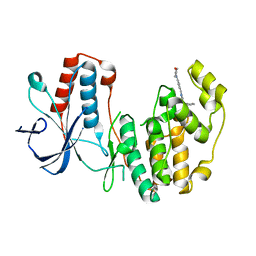 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9m | | Descriptor: | 2-(4-morpholin-4-ylphenyl)-~{N}4-(2-phenylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N67
 
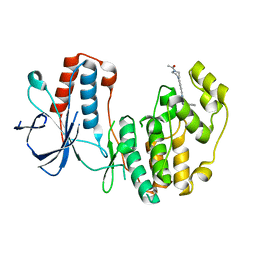 | |
5OMG
 
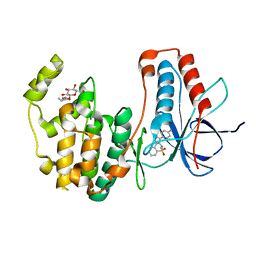 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXP4M12 | | Descriptor: | 3-(4-fluorophenyl)-4-methyl-1~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
8Z2C
 
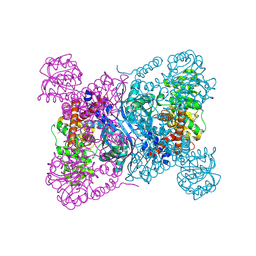 | |
8Z2X
 
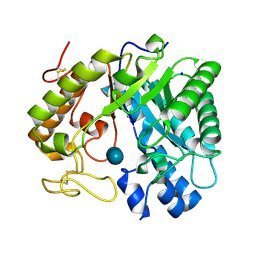 | | Crystal structure of exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) as a complex with cellobiose | | Descriptor: | Glucan 1,3-beta-glucosidase A, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 599, 2025
|
|
8Z2W
 
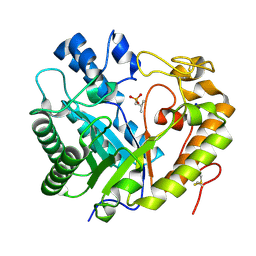 | | Crystal structure of apo- exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) | | Descriptor: | 2-methylpropylphosphonic acid, Glucan 1,3-beta-glucosidase A, SODIUM ION | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 599, 2025
|
|
5OMH
 
 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXH11 | | Descriptor: | 1-(3-chlorophenyl)-3-methyl-4~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
5N66
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9j | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 14, ~{N}4-[[4-(cyclopropylmethyl)furan-2-yl]methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
8YTD
 
 | | Crystal Structure of TrkA D5 domain in complex with two different macrocyclic peptides | | Descriptor: | 1,2-ETHANEDIOL, High affinity nerve growth factor receptor, Macrocyclic Peptide | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
8YTE
 
 | | Crystal Structure of TrkA D5 domain in complex with macrocyclic peptide | | Descriptor: | 1,2-ETHANEDIOL, AMINOMETHYLAMIDE, High affinity nerve growth factor receptor, ... | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
8Z0G
 
 | | Crystal structure of NeIle complexed with isoleucine | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Gabdukhakov, A, Subach, F.V. | | Deposit date: | 2024-04-09 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NeIle, a Genetically Encoded Indicator for Branched-Chain Amino Acids Based on mNeonGreen Fluorescent Protein and LIVBP Protein.
ACS Sens, 9, 2024
|
|
2JE2
 
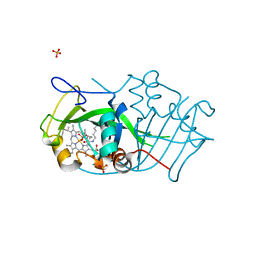 | | Cytochrome P460 from Nitrosomonas europaea - probable nonphysiological oxidized form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
2JE3
 
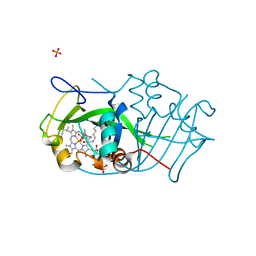 | | Cytochrome P460 from Nitrosomonas europaea - probable physiological form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
3R2Y
 
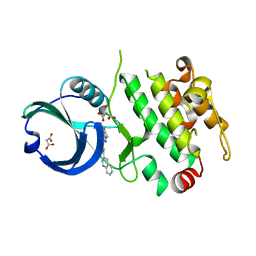 | | MK2 kinase bound to Compound 1 | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MALONATE ION, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Leonard, P. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8ZFV
 
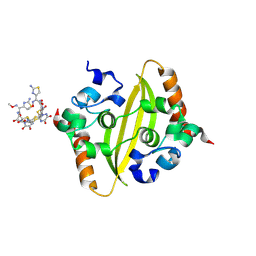 | | Crystal Structure of C-terminal domain of nucleocapsid protein from SARS-CoV-2 in complex with ceftriaxone | | Descriptor: | Ceftriaxone, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Dhaka, P, Mahto, J.K, Tomar, S, Kumar, P. | | Deposit date: | 2024-05-08 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the RNA binding inhibitors of the C-terminal domain of the SARS-CoV-2 nucleocapsid.
J.Struct.Biol., 217, 2025
|
|
5N63
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9c | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}4-[(4-fluorophenyl)methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
3R1N
 
 | | MK3 kinase bound to Compound 5b | | Descriptor: | 2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one, MAP kinase-activated protein kinase 3 | | Authors: | Oubrie, A, Kazemier, B. | | Deposit date: | 2011-03-11 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R30
 
 | | MK2 kinase bound to Compound 2 | | Descriptor: | 1-(2-aminoethyl)-3-[2-(quinolin-3-yl)pyridin-4-yl]-1H-pyrazole-5-carboxylic acid, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Fisher, M. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R2B
 
 | | MK2 kinase bound to Compound 5b | | Descriptor: | 2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, van Zeeland, M, Versteegh, J. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
