6VRU
 
 | | PIM-inhibitor complex 1 | | Descriptor: | 3,4-dichloro-2-cyclopropyl-1-[(piperidin-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Barberis, C.E, Batchelor, J.D, Mechin, I, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4A01
 
 | | Crystal Structure of the H-Translocating Pyrophosphatase | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, IMIDODIPHOSPHORIC ACID, MAGNESIUM ION, ... | | Authors: | Lin, S.-M, Tsai, J.-Y, Hsiao, C.-D, Chiu, C.-L, Pan, R.-L, Sun, Y.-J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-28 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of a Membrane Embedded H1-Translocating Pyrophosphatase
Nature, 484, 2012
|
|
6OC4
 
 | | CSP1-cyc(Dab6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OBW
 
 | | CSP1-cyc(K6D10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8OKU
 
 | | Salt-Inducible Kinase 3 in complex with an inhibitor | | Descriptor: | Serine/threonine-protein kinase SIK3, ~{N}-ethyl-4-[5-[1-(2-hydroxyethyl)pyrazol-4-yl]benzimidazol-1-yl]-2,6-dimethoxy-benzamide | | Authors: | Flower, T.G, Leonard, P.M, Lamers, M.B.A.C, Mollat, P. | | Deposit date: | 2023-03-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Optimization of Selectivity and Pharmacokinetic Properties of Salt-Inducible Kinase Inhibitors that Led to the Discovery of Pan-SIK Inhibitor GLPG3312.
J.Med.Chem., 67, 2024
|
|
1VRO
 
 | | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure | | Descriptor: | 5'-D(*CP*(GMS)P*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Wilds, C.J, Pattanayek, R, Pan, C, Wawrzak, Z, Egli, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure
J.Am.Chem.Soc., 124, 2002
|
|
1WUG
 
 | | complex structure of PCAF bromodomain with small chemical ligand NP1 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-4-METHYL-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-07 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1WUM
 
 | | Complex structure of PCAF bromodomain with small chemical ligand NP2 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1XUT
 
 | | Solution structure of TACI-CRD2 | | Descriptor: | Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding.
J.Biol.Chem., 280, 2005
|
|
1XU1
 
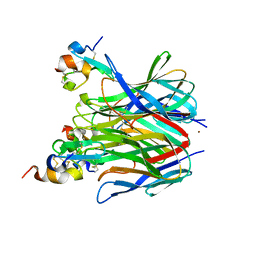 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1XUX
 
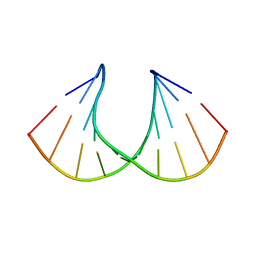 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMS)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
1XT9
 
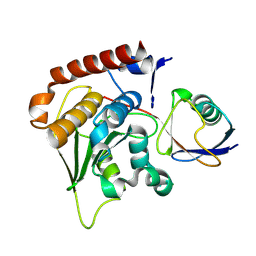 | | Crystal Structure of Den1 in complex with Nedd8 | | Descriptor: | Neddylin, Sentrin-specific protease 8 | | Authors: | Reverter, D, Wu, K, Erdene, T.G, Pan, Z.Q, Wilkinson, K.D, Lima, C.D. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Complex between Nedd8 and the Ulp/Senp Protease Family Member Den1.
J.Mol.Biol., 345, 2005
|
|
1XU2
 
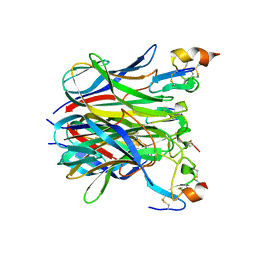 | | The crystal structure of APRIL bound to BCMA | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1XUW
 
 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMT)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
1YAU
 
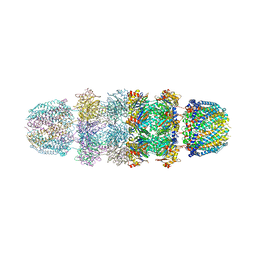 | | Structure of Archeabacterial 20S proteasome- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YAR
 
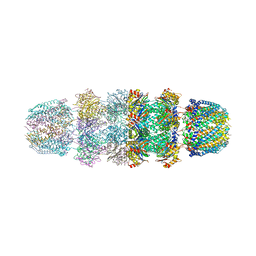 | | Structure of Archeabacterial 20S proteasome mutant D9S- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
8U37
 
 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with NVP-CJL037 at 2.48-A resolution | | Descriptor: | (6M)-3-amino-N-{4-[(3R,4S)-4-amino-3-methoxypiperidin-1-yl]pyridin-3-yl}-6-[3-(trifluoromethoxy)pyridin-2-yl]pyrazine-2-carboxamide, MAGNESIUM ION, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8UAK
 
 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8W6K
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BL03HB: Laue crystallography beamline at SSRF
To Be Published
|
|
8WSW
 
 | | The Crystal Structure of LIMK2a from Biortus | | Descriptor: | 1,2-ETHANEDIOL, LIM domain kinase 2, ~{N}-[5-[2-[2,6-bis(chloranyl)phenyl]-5-[bis(fluoranyl)methyl]pyrazol-3-yl]-1,3-thiazol-2-yl]-2-methyl-propanamide | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of LIMK2a from Biortus.
To Be Published
|
|
1YU9
 
 | | GppNHp-Bound Rab4A | | Descriptor: | GTP-binding protein, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
8X2T
 
 | |
8X23
 
 | | The Crystal Structure of MAPK13 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mitogen-activated protein kinase 13 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-09 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of MAPK13 from Biortus.
To Be Published
|
|
8X5L
 
 | | The Crystal Structure of PRKACA from Biortus. | | Descriptor: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, SODIUM ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of PRKACA from Biortus.
To Be Published
|
|
8X2A
 
 | | The Crystal Structure of BMX from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3S)-3-{[(2E)-but-2-enoyl]amino}piperidin-1-yl]-5-fluoro-2,3-dimethyl-1H-indole-7-carboxamide, CHLORIDE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-09 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Crystal Structure of BMX from Biortus.
To Be Published
|
|
