4OO0
 
 | |
7GGS
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0e5afe9d-1 (Mpro-x12696) | | Descriptor: | (4R)-6-chloro-N-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-c]pyridin-7-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5YXZ
 
 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | Authors: | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484
To Be Published
|
|
5YY1
 
 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-[2-(trifluoromethyl)phenyl]quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | Authors: | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739
To Be Published
|
|
2XOG
 
 | | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A | | Descriptor: | (2S,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-N-[[(2S,3S,4R,5R)-5-(2,4-DIOXOPYRIMIDIN-1-YL)-4-HYDROXY-2-(HYDROXYMETHYL)OXOLAN-3-YL]METHYLSULFONYL]-3,4-DIHYDROXY-OXOLANE-2-CARBOXAMIDE, RIBONUCLEASE PANCREATIC | | Authors: | Thiyagarajan, N, Smith, B.D, Raines, R.T, Acharya, K.R. | | Deposit date: | 2010-08-16 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A.
FEBS J., 278, 2011
|
|
1PIG
 
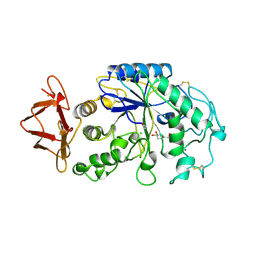 | | PIG PANCREATIC ALPHA-AMYLASE COMPLEXED WITH THE OLIGOSACCHARIDE V-1532 | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Machius, M, Vertesy, L, Huber, R, Wiegand, G. | | Deposit date: | 1996-06-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carbohydrate and protein-based inhibitors of porcine pancreatic alpha-amylase: structure analysis and comparison of their binding characteristics.
J.Mol.Biol., 260, 1996
|
|
4ZY0
 
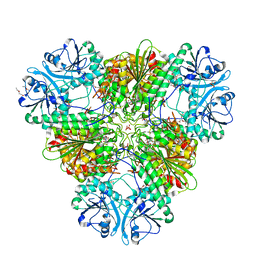 | |
7ZTV
 
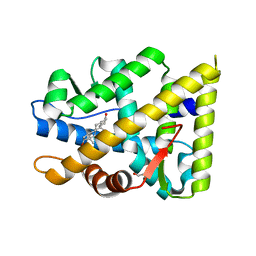 | | Crystal structure of mutant AR-LBD (F755L) bound to dihydrotestosterone | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
6LZ0
 
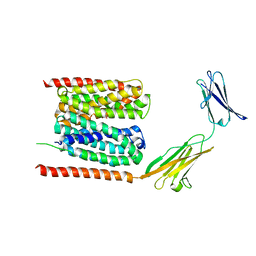 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7GGI
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-14ad9fe9-1 (Mpro-x12640) | | Descriptor: | 2-(3-chlorophenyl)-N-(1,2,3,4-tetrahydro-1,7-naphthyridin-5-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5ANK
 
 | | Crystal structure of CDK2 in complex with 2,4,6-trioxo-1-phenyl- hexahydropyrimidine-5-carboxamide processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 2,4,6-TRIOXO-1-PHENYL-HEXAHYDROPYRIMIDINE-5-CARBOXAMIDE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
6LUK
 
 | |
5AZC
 
 | |
4ZZQ
 
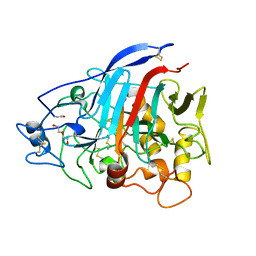 | | Dictyostelium discoideum cellobiohydrolase Cel7A apo structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, DI(HYDROXYETHYL)ETHER | | Authors: | Momeni, M.H, Hobdey, S.E, Knott, B, Beckham, G.T, Stahlberg, J. | | Deposit date: | 2015-04-13 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of Two Dictyostelium Cellobiohydrolases from the Amoebozoa Kingdom Reveal a High Conservation between Distant Phylogenetic Trees of Life.
Appl.Environ.Microbiol., 82, 2016
|
|
3L2U
 
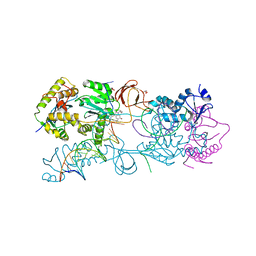 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and GS9137 (Elvitegravir) | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', 6-(3-chloro-2-fluorobenzyl)-1-[(1S)-1-(hydroxymethyl)-2-methylpropyl]-7-methoxy-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
6G9C
 
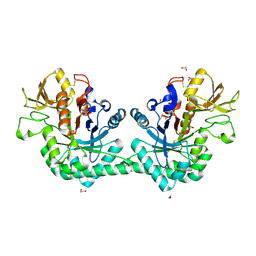 | | Crystal structure of immunomodulatory active chitinase from Trichuris suis, TsES1 | | Descriptor: | 1,2-ETHANEDIOL, Immunomodulatory active chitinase | | Authors: | Malecki, P.H, Balster, K, Hartmann, S, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Helminth-Derived Chitinase Structurally Similar to Mammalian Chitinase Displays Immunomodulatory Properties in Inflammatory Lung Disease.
J Immunol Res, 2021, 2021
|
|
1ZIV
 
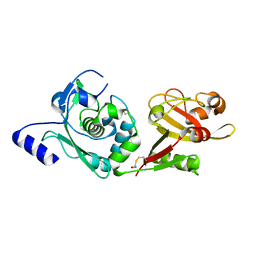 | | Catalytic Domain of Human Calpain-9 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain 9 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Dong, A, Choe, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition.
J.Mol.Biol., 366, 2007
|
|
6LVD
 
 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5ZHS
 
 | | Crystal structure of OsD14 in complex with covalently bound KK052 | | Descriptor: | (4-phenylpiperazin-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
7GF9
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TAT-ENA-80bfd3e5-4 (Mpro-x11590) | | Descriptor: | 1-[(4S)-3-(4-fluorobenzoyl)-2-methylindolizin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDB
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-968e8d9c-1 (Mpro-x10906) | | Descriptor: | (4S)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6GAR
 
 | | Crystal structure of oxidised ferredoxin/flavodoxin NADP+ oxidoreductase 1 (FNR1) from Bacillus cereus | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Skramo, S, Gudim, I, Hersleth, H.-P. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Characterization of Different Flavodoxin Reductase-Flavodoxin (FNR-Fld) Interactions Reveals an Efficient FNR-Fld Redox Pair and Identifies a Novel FNR Subclass.
Biochemistry, 57, 2018
|
|
5AM1
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 4-oxidanylidene-N-[(1S,2R)-2-phenylcyclopropyl]spiro[3H-chromene-2,4'-piperidine]-1'-carboxamide, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
6MAE
 
 | |
1ZL7
 
 | | Crystal structure of catalytically-active phospholipase A2 with bound calcium | | Descriptor: | CALCIUM ION, GLYCEROL, hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
