2BFV
 
 | |
2BFX
 
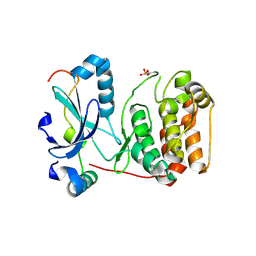 | | Mechanism of Aurora-B activation by INCENP and inhibition by Hesperadin. | | Descriptor: | AURORA KINASE B-A, INNER CENTROMERE PROTEIN A | | Authors: | Sessa, F, Mapelli, M, Ciferri, C, Tarricone, C, Areces, L.B, Schneider, T.R, Stukenberg, P.T, Musacchio, A. | | Deposit date: | 2004-12-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Aurora B Activation by Incenp and Inhibition by Hesperadin
Mol.Cell, 18, 2005
|
|
2BG9
 
 | |
2BXU
 
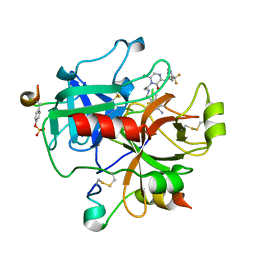 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | 1-(2-{[(6-AMINO-2-METHYLPYRIDIN-3-YL)METHYL]AMINO}ETHYL)-6-CHLORO-3-[(2,2-DIFLUORO-2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL, ALPHA THROMBIN, HIRUDIN | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-27 | | Release date: | 2006-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
2BGZ
 
 | | ATOMIC MODEL OF THE BACTERIAL FLAGELLAR BASED ON DOCKING AN X-RAY DERIVED HOOK STRUCTURE INTO AN EM MAP. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, Derosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BI6
 
 | | NMR STUDY OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
2BJX
 
 | | PROTEIN DISULFIDE ISOMERASE | | Descriptor: | PROTEIN (PROTEIN DISULFIDE ISOMERASE) | | Authors: | Kemmink, J, Dijkstra, K, Mariani, M, Scheek, R.M, Penka, E, Nilges, M, Darby, N.J. | | Deposit date: | 1999-01-30 | | Release date: | 1999-02-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure in solution of the b domain of protein disulfide isomerase.
J.Biomol.NMR, 13, 1999
|
|
8QFX
 
 | | Human Angiotensin-1 converting enzyme N-domain in complex with the lactotripeptide IPP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gregory, K.S, Acharya, K.R, Cozier, G.E. | | Deposit date: | 2023-09-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the inhibitory mechanism of angiotensin-I-converting enzyme by the lactotripeptides IPP and VPP.
Febs Lett., 598, 2024
|
|
8QHL
 
 | | Human Angiotensin-1 converting enzyme N-domain in complex with the lactotripeptide VPP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Gregory, K.S, Cozier, G.E, Acharya, K.R. | | Deposit date: | 2023-09-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the inhibitory mechanism of angiotensin-I-converting enzyme by the lactotripeptides IPP and VPP.
Febs Lett., 598, 2024
|
|
2BL6
 
 | | Solution structure of the Zn complex of EIAV NCp11(22-58) peptide, including two CCHC Zn-binding motifs. | | Descriptor: | NUCLEOCAPSID PROTEIN P11, ZINC ION | | Authors: | Amodeo, P, Castiglione-Morelli, M.A, Ostuni, A, Bavoso, A. | | Deposit date: | 2005-03-02 | | Release date: | 2006-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Features in Eiav Ncp11: A Lentivirus Nucleocapsid Protein with a Short Linker
Biochemistry, 45, 2006
|
|
2BNW
 
 | | Structural basis for cooperative binding of Ribbon-Helix-Helix Omega repressor to direct DNA heptad repeats | | Descriptor: | 5'-D(*CP*TP*TP*GP*TP*GP*AP*TP*TP*TP *GP*TP*GP*AP*TP*TP*CP*G)-3', 5'-D(*GP*AP*AP*TP*CP*AP*CP*AP*AP*AP *TP*CP*AP*CP*AP*AP*GP*C)-3', ORF OMEGA | | Authors: | Weihofen, W.A, Cicek, A, Pratto, F, Alonso, J.C, Saenger, W. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Omega Repressors Bound to Direct and Inverted DNA Repeats Explain Modulation of Transcription.
Nucleic Acids Res., 34, 2006
|
|
2C45
 
 | |
6CTW
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CCL2, beta, gamma dCTP analogue | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 4-amino-1-{2-deoxy-5-O-[(R)-{[(R)-[dichloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-L-threo-pentofuranosyl}pyrimidin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
2BSE
 
 | | Structure of Lactococcal Bacteriophage p2 Receptor Binding Protein in complex with a llama VHH domain | | Descriptor: | LLAMA IMMUNOGLOBULIN, RECEPTOR BINDING PROTEIN | | Authors: | Spinelli, S, Desmyter, A, Verrips, C.T, de Haard, H.J.W, Moineau, S, Cambillau, C. | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactococcal Bacteriophage P2 Receptor Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2BVD
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | (3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)piperidin-3-yl beta-D-glucopyranoside, ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-27 | | Release date: | 2005-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
2BVN
 
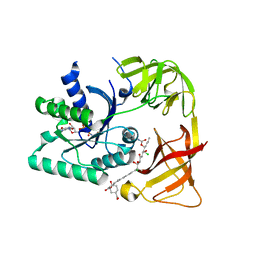 | | E. coli EF-Tu:GDPNP in complex with the antibiotic enacyloxin IIa | | Descriptor: | ELONGATION FACTOR TU, ENACYLOXIN IIA, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Watanabe, T, Nielsen, R.C, Dahlberg, C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-06-30 | | Release date: | 2005-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enacyloxin Iia Pinpoints a Binding Pocket of Elongation Factor TU for Development of Novel Antibiotics.
J.Biol.Chem., 281, 2006
|
|
2BLC
 
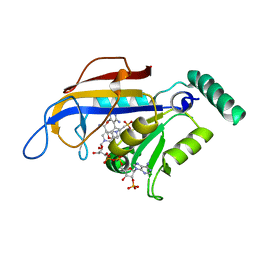 | | SP21 double mutant P. vivax Dihydrofolate reductase in complex with des-chloropyrimethamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-ETHYL-5-PHENYLPYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6ZSK
 
 | |
2BH5
 
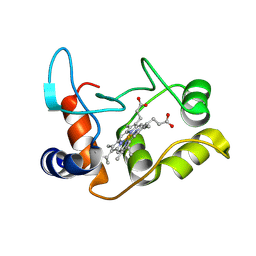 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 295 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
2BDS
 
 | |
2BE7
 
 | | Crystal structure of the unliganded (T-state) aspartate transcarbamoylase of the psychrophilic bacterium Moritella profunda | | Descriptor: | Aspartate Carbamoyltransferase Catalytic Chain, Aspartate Carbamoyltransferase Regulatory Chain, SULFATE ION, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella profunda.
J.Mol.Biol., 365, 2007
|
|
2BF8
 
 | | Crystal structure of SUMO modified ubiquitin conjugating enzyme E2- 25K | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-25 KDA, UBIQUITIN-LIKE PROTEIN SMT3C | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
6CR5
 
 | |
2C3D
 
 | | 2.15 Angstrom crystal structure of 2-ketopropyl coenzyme M oxidoreductase carboxylase with a coenzyme M disulfide bound at the active site | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
2BEV
 
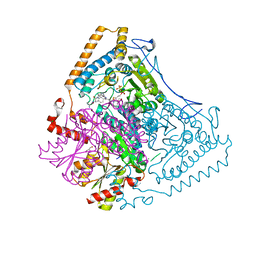 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-2-METHYL-BUTYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
