3J6G
 
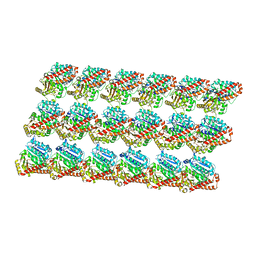 | | Minimized average structure of microtubules stabilized by taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
5XMJ
 
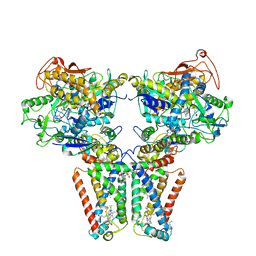 | | Crystal structure of quinol:fumarate reductase from Desulfovibrio gigas | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Guan, H.H, Hsieh, Y.C, Lin, P.R, Chen, C.J. | | Deposit date: | 2017-05-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Sci Rep, 8, 2018
|
|
6ZNP
 
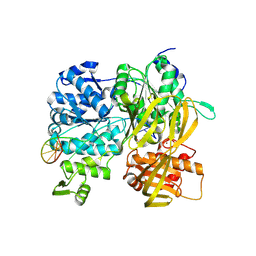 | | Crystal Structure of DUF1998 helicase MrfA bound to DNA | | Descriptor: | CITRIC ACID, Uncharacterized ATP-dependent helicase YprA, ZINC ION, ... | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
4DJB
 
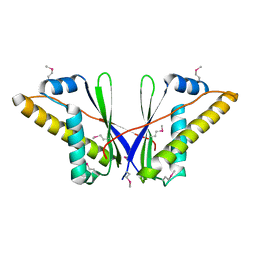 | | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors | | Descriptor: | E4-ORF3 | | Authors: | Ou, H.D, Kwiatkowski, W, Deerinck, T.J, Noske, A, Blain, K.Y, Land, H.S, Soria, C, Powers, C.J, May, A.P, Shu, X, Tsien, R.Y, Fitzpatrick, J.A.J, Long, J.A, Ellisman, M.H, Choe, S, O'Shea, C.C. | | Deposit date: | 2012-02-01 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors.
Cell(Cambridge,Mass.), 151, 2012
|
|
6Z7J
 
 | |
6Z7H
 
 | |
6Z7I
 
 | | Crystal structure of CTX-M-15 E166Q mutant apoenzyme | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 2022
|
|
6Z7K
 
 | | Crystal structure of CTX-M-15 in complex with the imine form of hydrolysed tazobactam | | Descriptor: | (2~{S},3~{S})-3-[bis(oxidanylidene)-$l^{5}-sulfanyl]-3-methyl-2-[(~{E})-3-oxidanylidenepropylideneamino]-4-(1,2,3-triaz ol-1-yl)butanoic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 2022
|
|
3JTO
 
 | | Crystal structure of the c-terminal domain of YpbH | | Descriptor: | Adapter protein mecA 2 | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the MecA Degradation Tag
To be Published
|
|
4USX
 
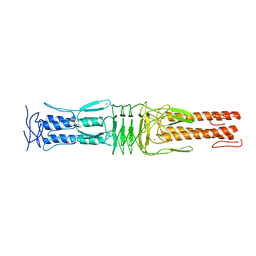 | | The Structure of the C-terminal YadA-like domain of BPSL2063 from Burkholderia pseudomallei | | Descriptor: | MAGNESIUM ION, TRIMERIC AUTOTRANSPORTER ADHESIN | | Authors: | Perletti, L, Gourlay, L.J, Peano, C, Pietrelli, A, DeBellis, G, Deantonio, C, Santoro, C, Sblattero, D, Bolognesi, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selecting Soluble/Foldable Protein Domains Through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6OGY
 
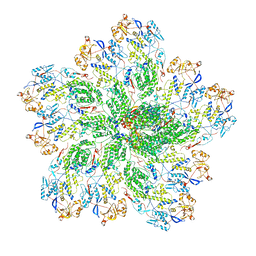 | | In situ structure of Rotavirus RNA-dependent RNA polymerase at duplex-open state | | Descriptor: | DNA/RNA (5'-D(*(GTG))-R(P*GP*C)-3'), Inner capsid protein VP2, RNA (5'-R(P*AP*GP*CP*C)-3'), ... | | Authors: | Ding, K, Chang, T, Shen, W, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release.
Nat Commun, 10, 2019
|
|
6ZXK
 
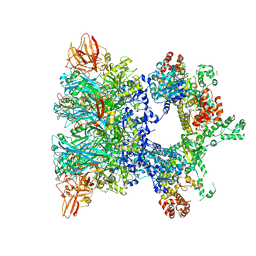 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state and PA7LF(2+1B) arrangement | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
8E23
 
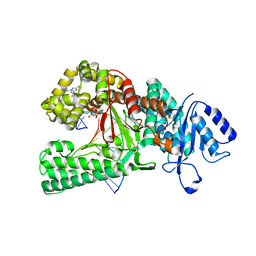 | | Human DNA polymerase theta in complex with allosteric inhibitor | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*GP*TP*CP*CP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*C)-3'), DNA (5'-D(*GP*C*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*G)-3'), ... | | Authors: | Mader, P, Pau, V.P.T, Sicheri, F. | | Deposit date: | 2022-08-13 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Identification of RP-6685 , an Orally Bioavailable Compound that Inhibits the DNA Polymerase Activity of Pol theta.
J.Med.Chem., 65, 2022
|
|
3J2V
 
 | | CryoEM structure of HBV core | | Descriptor: | PreC/core protein | | Authors: | Yu, X, Jin, L, Jih, J, Shih, C, Zhou, Z.H. | | Deposit date: | 2013-01-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | 3.5 angstrom cryoEM Structure of Hepatitis B Virus Core Assembled from Full-Length Core Protein.
Plos One, 8, 2013
|
|
7MI9
 
 | | Full integration complex of Cas1/Cas2 from Cas4-containing system | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (5'-D(P*CP*GP*GP*AP*AP*AP*AP*GP*AP*GP*CP*C)-3'), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
5Y4R
 
 | | Structure of a methyltransferase complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608, ... | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
6NU4
 
 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
7M60
 
 | | Structure of Mouse Importin alpha MLH1-S467A NLS Peptide Complex | | Descriptor: | DNA mismatch repair protein Mlh1 NLS peptide, Importin subunit alpha-1 | | Authors: | De Oliveira, H.C, Da Silva, T.D, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
7MJV
 
 | | MiaB in the complex with s-adenosylmethionine and RNA | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
4V0C
 
 | |
7MIB
 
 | |
6YVJ
 
 | | EED in complex with a triazolopyrimidine | | Descriptor: | GLYCEROL, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Free energy perturbation in the design of EED ligands as inhibitors of polycomb repressive complex 2 (PRC2) methyltransferase.
Bioorg.Med.Chem.Lett., 39, 2021
|
|
6YVI
 
 | | EED in complex with a cyano-benzofuran | | Descriptor: | 5-fluoranyl-4-[[[8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-yl]amino]methyl]-2,3-dihydro-1-benzofuran-7-carbonitrile, CALCIUM ION, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Free energy perturbation in the design of EED ligands as inhibitors of polycomb repressive complex 2 (PRC2) methyltransferase.
Bioorg.Med.Chem.Lett., 39, 2021
|
|
7MJZ
 
 | | The structure of MiaB with pentasulfide bridge | | Descriptor: | IRON/SULFUR CLUSTER, PENTASULFIDE-SULFUR, SODIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
5YEQ
 
 | | The structure of Sac-KARI protein | | Descriptor: | 1,2-ETHANEDIOL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Ko, T.P, Chen, C.Y, Lin, K.F, Lin, B.L, Huang, C.H, Chiang, C.H, Horng, J.C, Tsai, M.D. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NADH/NADPH bi-cofactor-utilizing and thermoactive ketol-acid reductoisomerase from Sulfolobus acidocaldarius
Sci Rep, 8, 2018
|
|
