6ALV
 
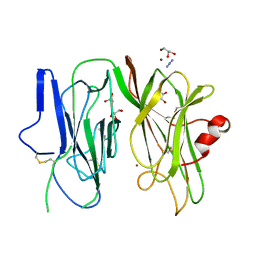 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
4NZ7
 
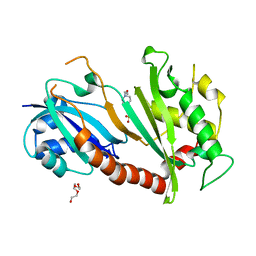 | | Steroid receptor RNA Activator (SRA) modification by the human Pseudouridine Synthase 1 (hPus1p): RNA binding, activity, and atomic model | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TRIETHYLENE GLYCOL, tRNA pseudouridine synthase A, ... | | Authors: | Huet, T, Thore, S. | | Deposit date: | 2013-12-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Steroid Receptor RNA Activator (SRA) Modification by the Human Pseudouridine Synthase 1 (hPus1p): RNA Binding, Activity, and Atomic Model
Plos One, 9, 2014
|
|
5TH4
 
 | |
5FXS
 
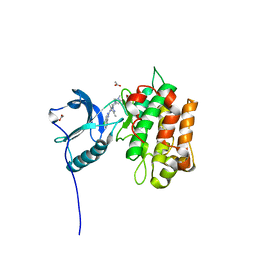 | | IGFR-1R complex with a pyrimidine inhibitor. | | Descriptor: | 2-[4-[4-[[(6Z)-5-chloranyl-6-pyrazolo[1,5-a]pyridin-3-ylidene-1H-pyrimidin-2-yl]amino]-3,5-dimethyl-pyrazol-1-yl]piperidin-1-yl]-N,N-dimethyl-ethanamide, ACETATE ION, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR | | Authors: | Degorce, S, Boyd, S, Curwen, J, Ducray, R, Halsall, C, Jones, C, Lach, F, Lenz, E, Pass, M, Pass, S, Trigwell, C, Norman, R, Phillips, C. | | Deposit date: | 2016-03-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent, Selective, Orally Bioavailable, and Efficacious Novel 2-(Pyrazol-4-ylamino)-pyrimidine Inhibitor of the Insulin-like Growth Factor-1 Receptor (IGF-1R).
J. Med. Chem., 59, 2016
|
|
1Z9T
 
 | |
3HSW
 
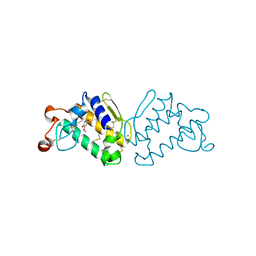 | | Crystal Structure of Porcine Pancreatic Phospholipase A2 in Complex with 2-methoxycyclohexa-2-5-diene-1,4-dione | | Descriptor: | 2-methoxycyclohexa-2,5-diene-1,4-dione, CALCIUM ION, Phospholipase A2, ... | | Authors: | Dileep, K.V, Tintu, I, Karthe, P, Mandal, P.K, Haridas, M, Sadasivan, C. | | Deposit date: | 2009-06-11 | | Release date: | 2009-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of porcine pancreatic phospholipase A2 in complex with 2-methoxycyclohexa-2-5-diene-1,4-dione
Frontiers in life sci., 2012
|
|
5BWX
 
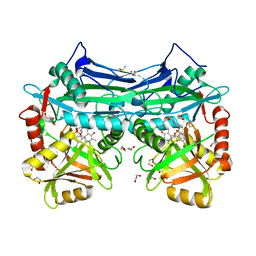 | |
4YHQ
 
 | | Crystal structure of multidrug resistant clinical isolate PR20 with GRL-5010A | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-02-27 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substituted Bis-THF Protease Inhibitors with Improved Potency against Highly Resistant Mature HIV-1 Protease PR20.
J.Med.Chem., 58, 2015
|
|
3NUF
 
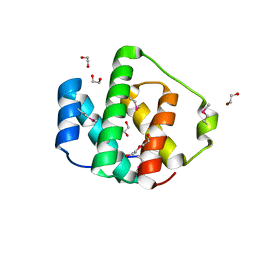 | |
6B4U
 
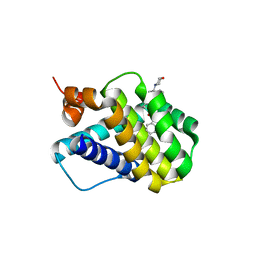 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(2-methylphenyl)-1-[2-(morpholin-4-yl)ethyl]-3-{3-[(naphthalen-1-yl)oxy]propyl}-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
3HVY
 
 | |
6B19
 
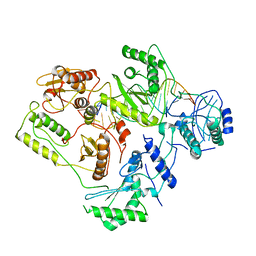 | | Architecture of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | RNA genome fragment, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit, ... | | Authors: | Larsen, K.P, Mathiharan, Y.K, Chen, D.H, Puglisi, J.D, Skiniotis, G, Puglisi, E.V. | | Deposit date: | 2017-09-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of an HIV-1 reverse transcriptase initiation complex.
Nature, 557, 2018
|
|
5GSW
 
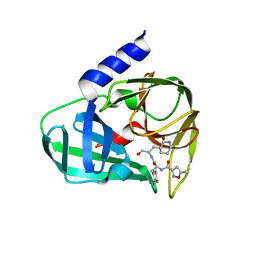 | | Crystal structure of EV71 3C in complex with N69S 1.8k | | Descriptor: | 3C protein, ~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]propan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wang, Y. | | Deposit date: | 2016-08-17 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the Enterovirus 71 3C Protease in Complex with NK-1.8k and Indications for the Development of Antienterovirus Protease Inhibitor
Antimicrob. Agents Chemother., 61, 2017
|
|
6TTC
 
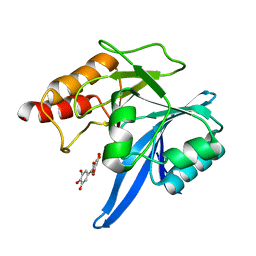 | | Haddock model of NDM-1/myricetin complex | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
7XHQ
 
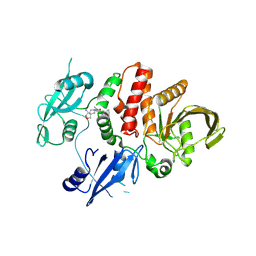 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[(3S,4R)-4-azanyl-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-04-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the SHP2 allosteric inhibitor 2-((3R,4R)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl)-5-(2,3-dichlorophenyl)-3-methylpyrrolo[2,1-f][1,2,4] triazin-4(3H)-one.
J Enzyme Inhib Med Chem, 38, 2023
|
|
5CR5
 
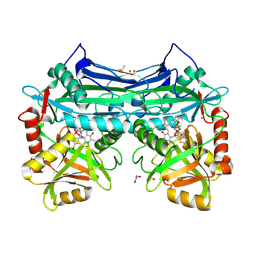 | |
3IEE
 
 | |
7C7X
 
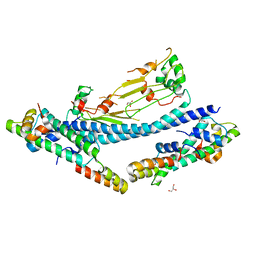 | |
4ZAD
 
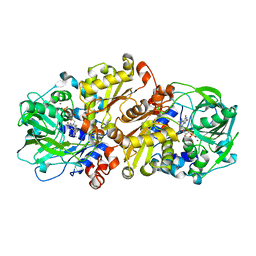 | | Structure of C. dubliensis Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
5GSO
 
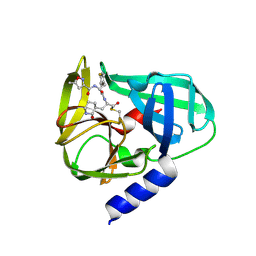 | | Crystal Structures of EV71 3C Protease in complex with NK-1.8k | | Descriptor: | 3C protein, ~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]propan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wang, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Enterovirus 71 3C Protease in Complex with NK-1.8k and Indications for the Development of Antienterovirus Protease Inhibitor
Antimicrob. Agents Chemother., 61, 2017
|
|
4Q0Y
 
 | |
8D9V
 
 | | gamma-Arf1 homodimeric interface within AP-1, Arf1, Nef lattice on narrow membrane tubes | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D9T
 
 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes, centered on gamma-Arf1 | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
4Q6K
 
 | |
8D9U
 
 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes, centered on beta-Arf1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
