3MWW
 
 | | Crystal structure of HCV NS5B polymerase | | Descriptor: | 1-[2-(4-carboxypiperidin-1-yl)-2-oxoethyl]-3-cyclohexyl-2-furan-3-yl-1H-indole-6-carboxylic acid, Genome polyprotein, SULFATE ION | | Authors: | Coulombe, R. | | Deposit date: | 2010-05-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
5H2Q
 
 | | Crystal structure of T brucei phosphodiesterase B2 bound to compound 13e | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[2-(2-oxidanylideneimidazolidin-1-yl)ethoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, MAGNESIUM ION, Phosphodiesterase, ... | | Authors: | Noble, C.G. | | Deposit date: | 2016-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Trypanosomal Phosphodiesterase B1 and B2 as a Potential Therapy for Human African Trypanosomiasis
To Be Published
|
|
5H35
 
 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
8PB1
 
 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1), obtained after local refinement. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-06-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4R8Y
 
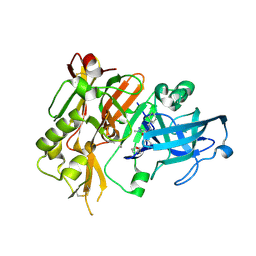 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4Z6H
 
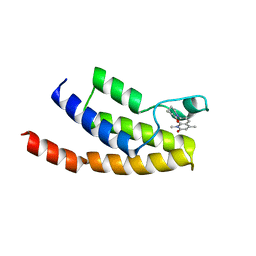 | | Crystal structure of BRD9 bromodomain in complex with a valerolactam quinolone ligand | | Descriptor: | 1,4-dimethyl-7-(2-oxopiperidin-1-yl)quinolin-2(1H)-one, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5DGT
 
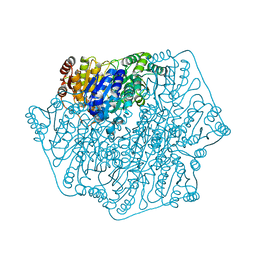 | | BENZOYLFORMATE DECARBOXYLASE H70A MUTANT at pH 8.5 FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Bera, A.K, Hasson, M.S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | BENZOYLFORMATE DECARBOXYLASE H70A MUTANT at pH 8.5 FROM PSEUDOMONAS PUTIDA
to be published
|
|
4RA7
 
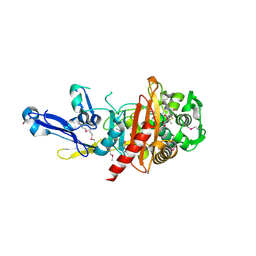 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-2-hydroxy-1-{[(2-hydroxynaphthalen-1-yl)carbonyl]amino}ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin
To be Published
|
|
6TCN
 
 | | Crystal structure of the omalizumab Fab - crystal form II | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6BCR
 
 | |
6Q4Z
 
 | | Structure of an inactive variant (D94N) of MPT-2, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana, in complex with beta-1,2-mannobiose | | Descriptor: | LmxM MPT-2 D94N, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Kloehn, J, Viera-Lara, M, Stanton, L, Cobbold, S, Pires, D.E, Hanssen, E, Parker, M.W, Ascher, D.B, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6B5M
 
 | |
5KAC
 
 | | Protein Tyrosine Phosphatase 1B P185G mutant, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Choy, M.S, Machado, L.E.S.F, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
9LSL
 
 | | The crystal structure of PDE5A with L1 | | Descriptor: | (13~{S},15~{R})-15-(3-chloranyl-4-methoxy-phenyl)-12-ethanoyl-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaene-4-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
4GKZ
 
 | | HA1.7, a MHC class II restricted TCR specific for haemagglutinin | | Descriptor: | 1,2-ETHANEDIOL, Alpha chain of Class II TCR, Beta Chain of Class II TCR, ... | | Authors: | Holland, C.J, Rizkallah, P.J, Cole, D.K, Sewell, A.K, Godkin, A.J. | | Deposit date: | 2012-08-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Minimal conformational plasticity enables TCR cross-reactivity to different MHC class II heterodimers.
Sci Rep, 2, 2012
|
|
6Y8F
 
 | | An inactive (D136N and D137N) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotriose | | Descriptor: | Alpha-1,6-endo-mannanase GH76A mutant, CALCIUM ION, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose | | Authors: | Hehemann, J.H, Solanki, V.A. | | Deposit date: | 2020-03-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Glycoside hydrolase from the GH76 family indicates that marine Salegentibacter sp. Hel_I_6 consumes alpha-mannan from fungi.
Isme J, 2022
|
|
4RFY
 
 | | Crystal structure of BTK kinase domain complexed with 6-(dimethylamino)-2-[2-(hydroxymethyl)-3-[1-methyl-5-[[5-(morpholine-4-carbonyl)-2-pyridyl]amino]-6-oxo-3-pyridyl]phenyl]-3,4-dihydroisoquinolin-1-one | | Descriptor: | 6-(dimethylamino)-2-[2-(hydroxymethyl)-3-(1-methyl-5-{[5-(morpholin-4-ylcarbonyl)pyridin-2-yl]amino}-6-oxo-1,6-dihydropyridin-3-yl)phenyl]-3,4-dihydroisoquinolin-1(2H)-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Finding the perfect spot for fluorine: Improving potency up to 40-fold during a rational fluorine scan of a Bruton's Tyrosine Kinase (BTK) inhibitor scaffold.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6Q76
 
 | |
11BG
 
 | | A POTENTIAL ALLOSTERIC SUBSITE GENERATED BY DOMAIN SWAPPING IN BOVINE SEMINAL RIBONUCLEASE | | Descriptor: | PROTEIN (BOVINE SEMINAL RIBONUCLEASE), SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Adinolfi, S, Sica, F, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-11 | | Release date: | 1999-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease.
J.Mol.Biol., 293, 1999
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
6F8W
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-18a | | Descriptor: | 3-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
4JQ6
 
 | | Crystal structure of blue light-absorbing proteorhodopsin from Med12 at 2.3 Angstrom | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Proteorhodopsin, RETINAL | | Authors: | Ozorowski, G, Luecke, H. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4QSS
 
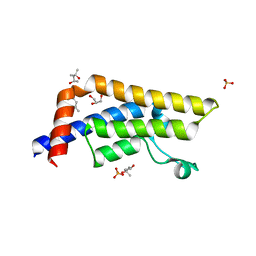 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-methylpyrrolidin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
7TQQ
 
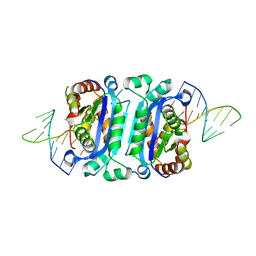 | | Structure of human TREX1-DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*GP*GP*CP*CP*GP*GP*CP*CP*AP*TP*C)-3'), Three-prime repair exonuclease 1 | | Authors: | Zhou, W, Richmond-Buccola, D, Kranzusch, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of human TREX1 DNA degradation and autoimmune disease.
Nat Commun, 13, 2022
|
|
8ZR4
 
 | | Cryo-EM structure of the N2-4N2C402 complex at a resolution of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4N2C402_Fab_H, 4N2C402_Fab_L, ... | | Authors: | Chu, B.X, Wang, X, Yang, Q, Wu, X.D, Zhang, Z.L. | | Deposit date: | 2024-06-03 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Profiling influenza neuraminidase pre-existing humoral immunity in humans
Cell Host Microbe, 2024
|
|
