7BG9
 
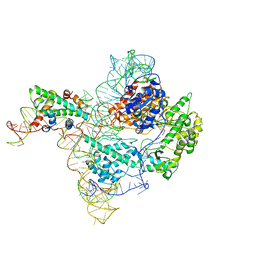 | | The catalytic core lobe of human telomerase in complex with a telomeric DNA substrate | | Descriptor: | DNA (5'-D(P*TP*TP*AP*GP*GP*G)-3'), Histone H2A, Histone H2B, ... | | Authors: | Nguyen, T.H.D, Ghanim, G.E, Fountain, A.J, van Roon, A.M.M, Rangan, R, Das, R, Collins, K. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of human telomerase holoenzyme with bound telomeric DNA.
Nature, 593, 2021
|
|
8GZG
 
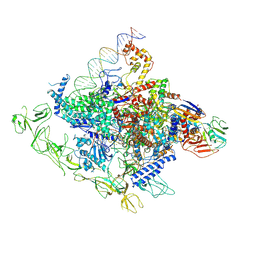 | | Cryo-EM structure of Synechocystis sp. PCC 6803 RPitc | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shen, L.Q, You, L.L, Zhang, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC 6803 CTP-bound RPitc
Proc.Natl.Acad.Sci.USA, 2023
|
|
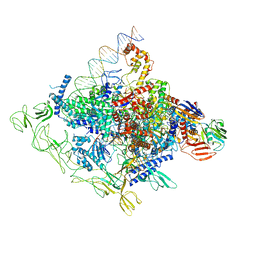
8GZH
 
 | |
6ZLW
 
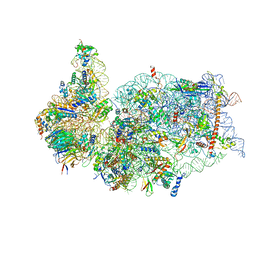 | | SARS-CoV-2 Nsp1 bound to the human 40S ribosomal subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6YBW
 
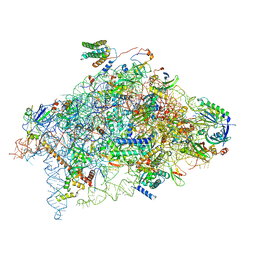 | | Structure of a human 48S translational initiation complex - 40S body | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6XE0
 
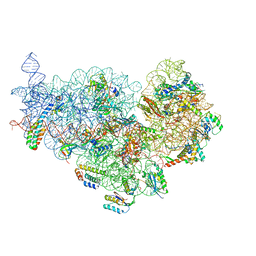 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
4Q5V
 
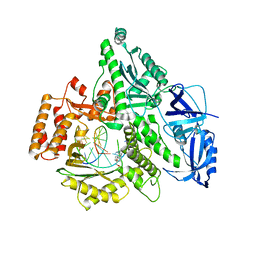 | | Crystal structure of the catalytic core of human DNA polymerase alpha in ternary complex with an RNA-primed DNA template and aphidicolin | | Descriptor: | (3R,4R,4aR,6aS,8R,9R,11aS,11bS)-4,9-bis(hydroxymethyl)-4,11b-dimethyltetradecahydro-8,11a-methanocyclohepta[a]naphthale ne-3,9-diol, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Baranovskiy, A.G, Babayeva, N.D, Suwa, Y, Gu, J, Tahirov, T.H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for inhibition of DNA replication by aphidicolin.
Nucleic Acids Res., 42, 2014
|
|
7MDZ
 
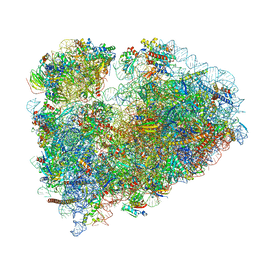 | | 80S rabbit ribosome stalled with benzamide-CHX | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S21, ... | | Authors: | Koga, Y, Hoang, E.M, Park, Y, Keszei, A.F.A, Murray, J, Shao, S, Liau, B.B. | | Deposit date: | 2021-04-06 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of C13-Aminobenzoyl Cycloheximide Derivatives that Potently Inhibit Translation Elongation.
J.Am.Chem.Soc., 143, 2021
|
|
7A5P
 
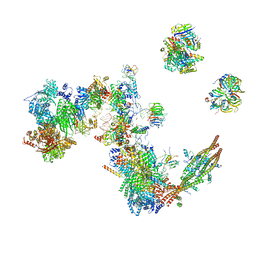 | | Human C Complex Spliceosome - Medium-resolution PERIPHERY | | Descriptor: | Cell division cycle 5-like protein, Crooked neck-like protein 1, Eukaryotic initiation factor 4A-III, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
6ZYM
 
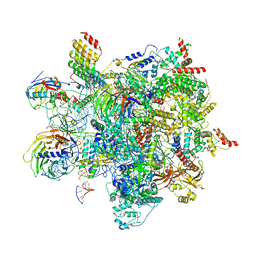 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
5O9G
 
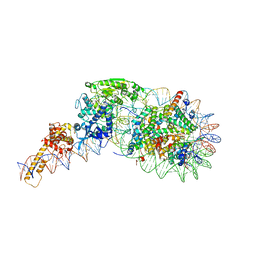 | | Structure of nucleosome-Chd1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
6G18
 
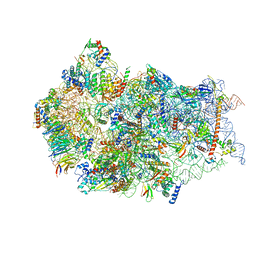 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State C | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G4S
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State B | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S13, 40S ribosomal protein S14, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
4TY9
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYB
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYA
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TY8
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 6-methyl-2H-chromen-2-one, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
7YE1
 
 | | The cryo-EM structure of C. crescentus GcrA-TACup | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (57-MER)-non template, DNA (57-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7YE2
 
 | | The cryo-EM structure of C. crescentus GcrA-TACdown | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (90-MER)-non template, DNA (90-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
5N65
 
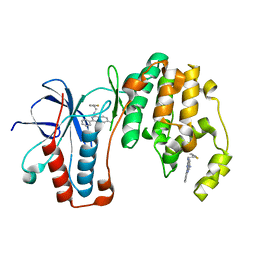 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9h | | Descriptor: | 2-phenyl-~{N}4-(2-thiophen-2-ylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N64
 
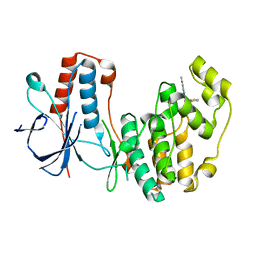 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9g | | Descriptor: | 2-phenyl-~{N}4-(thiophen-2-ylmethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5TW1
 
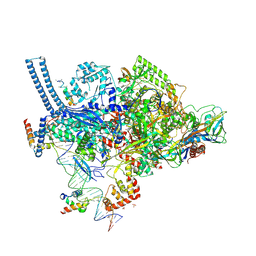 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with RbpA | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Hubin, E.A, Darst, S.A, Campbell, E.A. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA.
Elife, 6, 2017
|
|
5L3T
 
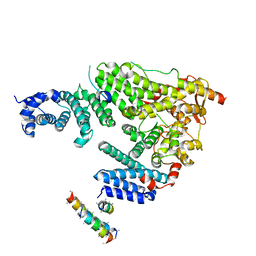 | | Structure of the Saccharomyces cerevisiae TREX-2 complex | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1, ... | | Authors: | Stewart, M, Aibara, S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.927 Å) | | Cite: | The Sac3 TPR-like region in the Saccharomyces cerevisiae TREX-2 complex is more extensive but independent of the CID region.
J. Struct. Biol., 195, 2016
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
