1KSG
 
 | | Complex of Arl2 and PDE delta, Crystal Form 1 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RETINAL ROD RHODOPSIN-SENSITIVE CGMP 3',5'-CYCLIC PHOSPHODIESTERASE DELTA-SUBUNIT, ... | | Authors: | Hanzal-Bayer, M, Renault, L, Roversi, P, Wittinghofer, A, Hillig, R.C. | | Deposit date: | 2002-01-13 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex of Arl2-GTP and PDE delta: from structure to function.
EMBO J., 21, 2002
|
|
8DLI
 
 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
4XX4
 
 | |
7TGP
 
 | | E176T/Y188G variant of the internal UBA Domain of HHR23A | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Rothfuss, M, Bowler, B.E, McClelland, L.J, Sprang, S.R. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
5FXM
 
 | |
7FZZ
 
 | | Crystal Structure of human FABP4 in complex with 3-[(2,4-dichloro-5-methylphenyl)sulfonylamino]thiophene-2-carboxamide, i.e. SMILES C1(=C(SC=C1)C(=O)N)NS(=O)(=O)c1c(cc(c(c1)C)Cl)Cl with IC50=0.199 microM | | Descriptor: | 3-[(2,4-dichloro-5-methylbenzene-1-sulfonyl)amino]thiophene-2-carboxamide, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5BU3
 
 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | Descriptor: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | Authors: | Pan, L, Guo, Y, Liu, J. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
4XAT
 
 | | Crystal structure of the olfactomedin domain from noelin/pancortin/olfactomedin-1 | | Descriptor: | CALCIUM ION, GLYCEROL, Noelin, ... | | Authors: | Hill, S.E, Nguyen, E, Lieberman, R.L. | | Deposit date: | 2014-12-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Molecular Details of Olfactomedin Domains Provide Pathway to Structure-Function Studies.
Plos One, 10, 2015
|
|
7FWX
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 | | Descriptor: | (4E)-6-(5-methoxy-3,6,7-trimethyl-1,2-benzoxazol-4-yl)-4-methylhex-4-enoic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8D4T
 
 | | Mammalian CIV with GDN bound | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, ... | | Authors: | Di Trani, J, Rubinstein, J. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of mammalian complex IV inhibition by steroids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3LL7
 
 | | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase | | Authors: | Chang, C, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative methyltransferase PG_1098 from Porphyromonas gingivalis W83
To be Published
|
|
7AD6
 
 | | Crystal structure of human complement C5 in complex with the K92 bovine knob domain peptide. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
7X5P
 
 | | Truncated VhChiP (1-19aa) in complex with doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, ... | | Authors: | Sanram, S, Robinson, C.R, Aunkham, A, Suginta, W. | | Deposit date: | 2022-03-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of truncated VhChiP
To Be Published
|
|
5FP6
 
 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol (AT17833) in an alternate binding site. | | Descriptor: | 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5I24
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF021 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-amino-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
6FQM
 
 | | 3.06A COMPLEX OF S.AUREUS GYRASE with imidazopyrazinone T1 AND DNA | | Descriptor: | 7-[(3~{S})-3-azanylpyrrolidin-1-yl]-5-cyclopropyl-8-fluoranyl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
5I2N
 
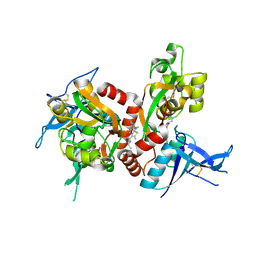 | | Structure of the human GluN1/GluN2A LBD in complex with N-ethyl-7-{[2-fluoro-3-(trifluoromethyl)phenyl]methyl}-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 29) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
6D56
 
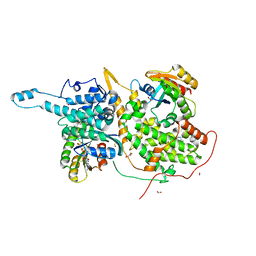 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-2-(2,6-diazaspiro[3.3]heptan-2-yl)-4-(3,5-dimethyl-1H-pyrazol-4-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
2RF0
 
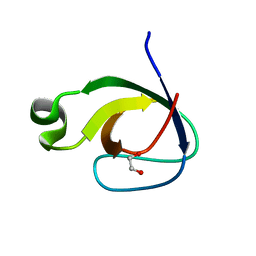 | | Crystal structure of human mixed lineage kinase MAP3K10 SH3 domain | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase 10 | | Authors: | Ugochukwu, E, Eswaran, J, Elkins, J, Keates, T, Pike, A.C.W, Berridge, G, Savitsky, P, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-27 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human Mixed lineage kinase MAP3K10 SH3 domain.
To be Published
|
|
6P68
 
 | |
8FYN
 
 | | MicroED structure of A2A from plasma milled lamellae | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
6RV1
 
 | | human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Mirco, D. | | Deposit date: | 2019-05-30 | | Release date: | 2020-04-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cycloserine enantiomers are reversible inhibitors of human alanine:glyoxylate aminotransferase: implications for Primary Hyperoxaluria type 1.
Biochem.J., 476, 2019
|
|
5B4B
 
 | | Crystal structure of LpxH with lipid X in spacegroup C2 | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
9CUB
 
 | | Human STING G230A/R293Q variant bound to diABZI-a1 | | Descriptor: | 1,1'-[(2E)-but-2-ene-1,4-diyl]bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-1,3-benzimidazole-5-carboxamide}, Stimulator of interferon genes protein | | Authors: | Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING.
Nat Commun, 16, 2025
|
|
6P6Y
 
 | | Crystal structure of voltage-gated sodium channel NavAb V100C/Q150C disulfide crosslinked mutant in the activated state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
