3IK8
 
 | |
3IKG
 
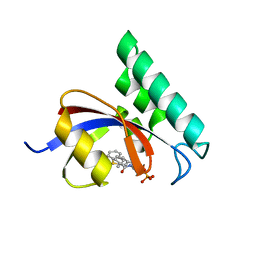 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | 分子名称: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | 登録日 | 2009-08-05 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3JYJ
 
 | |
3KAC
 
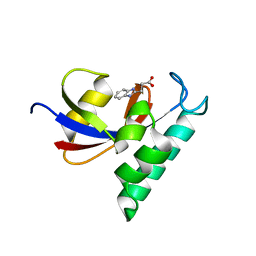 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | 分子名称: | 3-(1H-benzimidazol-2-yl)propanoic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | 登録日 | 2009-10-19 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1M5Y
 
 | |
2PV3
 
 | |
7SA5
 
 | | Two-state solution NMR structure of Apo Pin1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Born, A, Vogeli, B. | | 登録日 | 2021-09-22 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Reconstruction of Coupled Intra- and Interdomain Protein Motion from Nuclear and Electron Magnetic Resonance.
J.Am.Chem.Soc., 143, 2021
|
|
1F8A
 
 | | STRUCTURAL BASIS FOR THE PHOSPHOSERINE-PROLINE RECOGNITION BY GROUP IV WW DOMAINS | | 分子名称: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1, Y(SEP)PT(SEP)S PEPTIDE | | 著者 | Verdecia, M.A, Bowman, M.E, Lu, K.P, Hunter, T, Noel, J.P. | | 登録日 | 2000-06-29 | | 公開日 | 2000-08-23 | | 最終更新日 | 2013-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural basis for phosphoserine-proline recognition by group IV WW domains.
Nat.Struct.Biol., 7, 2000
|
|
4QIB
 
 | | Oxidation-Mediated Inhibition of the Peptidyl-Prolyl Isomerase Pin1 | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | 著者 | Innes, B.T, Sowole, M.A, Konermann, L, Litchfield, D.W, Brandl, C.J, Shilton, B.H. | | 登録日 | 2014-05-30 | | 公開日 | 2015-02-04 | | 最終更新日 | 2015-03-04 | | 実験手法 | X-RAY DIFFRACTION (1.865 Å) | | 主引用文献 | Peroxide-mediated oxidation and inhibition of the peptidyl-prolyl isomerase Pin1.
Biochim.Biophys.Acta, 1852, 2015
|
|
8HM3
 
 | | Complex of PPIase-BfUbb | | 分子名称: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | 著者 | Xu, J.H, Chen, Z, Gao, X. | | 登録日 | 2022-12-02 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
1YW5
 
 | | Peptidyl-prolyl isomerase ESS1 from Candida albicans | | 分子名称: | peptidyl prolyl cis/trans isomerase | | 著者 | Li, Z, Li, H, Devasahayam, G, Gemmill, T, Chaturvedi, V, Hanes, S.D, Van Roey, P. | | 登録日 | 2005-02-17 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Structure of the Candida albicans Ess1 Prolyl Isomerase Reveals a Well-Ordered Linker that Restricts Domain Mobility
Biochemistry, 44, 2005
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-06-05 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
1ZCN
 
 | | human Pin1 Ng mutant | | 分子名称: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | 登録日 | 2005-04-12 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6O34
 
 | |
6O33
 
 | |
6VAJ
 
 | | Crystal Structure Analysis of human PIN1 | | 分子名称: | 2-chloro-N-(2,2-dimethylpropyl)-N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2019-12-17 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo.
Nat.Chem.Biol., 17, 2021
|
|
7KKF
 
 | |
5UY9
 
 | | Prolyl isomerase Pin1 R14A mutant bound with Brd4 peptide | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Brd4 peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | 著者 | Dong, S.-H, Nair, S. | | 登録日 | 2017-02-23 | | 公開日 | 2017-04-26 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Prolyl isomerase PIN1 regulates the stability, transcriptional activity and oncogenic potential of BRD4.
Oncogene, 36, 2017
|
|
7SUQ
 
 | |
7SUR
 
 | |
1NMV
 
 | | Solution structure of human Pin1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | 登録日 | 2003-01-11 | | 公開日 | 2003-08-12 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
3NTP
 
 | | Human Pin1 complexed with reduced amide inhibitor | | 分子名称: | (2R)-2-(acetylamino)-3-[(2S)-2-{[2-(1H-indol-3-yl)ethyl]carbamoyl}pyrrolidin-1-yl]propyl dihydrogen phosphate, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Zhang, Y. | | 登録日 | 2010-07-05 | | 公開日 | 2012-01-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.762 Å) | | 主引用文献 | A reduced-amide inhibitor of Pin1 binds in a conformation resembling a twisted-amide transition state.
Biochemistry, 50, 2011
|
|
3ODK
 
 | | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-pyridin-2-yl-1H-pyrazole-5-carboxylic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | 登録日 | 2010-08-11 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OOB
 
 | | Structural and functional insights of directly targeting Pin1 by Epigallocatechin-3-gallate | | 分子名称: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | 著者 | Urusova, D.V, Shim, J.-H, Kim, D.-J, Jung, S.K, Zykova, T.A, Bode, A.M, Dong, Z. | | 登録日 | 2010-08-30 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1.
Cancer Prev Res (Phila), 4, 2011
|
|
1PIN
 
 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | 分子名称: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | 著者 | Noel, J.P, Ranganathan, R, Hunter, T. | | 登録日 | 1998-06-21 | | 公開日 | 1998-10-14 | | 最終更新日 | 2023-07-26 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
