3U8Z
 
 | | human merlin FERM domain | | Descriptor: | Merlin | | Authors: | Yogesha, S.D, Sharff, A.J, Giovannini, M, Bricogne, G, Izard, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Unfurling of the band 4.1, ezrin, radixin, moesin (FERM) domain of the merlin tumor suppressor.
Protein Sci., 20, 2011
|
|
3QIJ
 
 | | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R | | Descriptor: | Protein 4.1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Zhong, N, Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-27 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R
to be published
|
|
3ZDT
 
 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5MV7
 
 | | Structure of human Myosin 7b C-terminal MyTH4-FERM MF2 domain | | Descriptor: | SULFATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE, Unconventional myosin-VIIb | | Authors: | Planelles-Herrero, V.J, Sourigues, Y, Titus, M.A, Houdusse, A. | | Deposit date: | 2017-01-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Myosin 7 and its adaptors link cadherins to actin.
Nat Commun, 8, 2017
|
|
5MV9
 
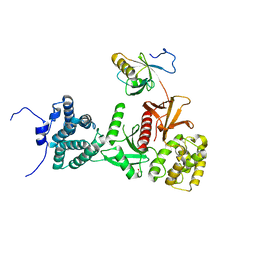 | | Structure of human Myosin 7a C-terminal MyTH4-FERM domain in complex with harmonin-a PDZ3 domain | | Descriptor: | ACETATE ION, Unconventional myosin-VIIa, cDNA FLJ51329, ... | | Authors: | Yu, I, Sourigues, Y, Titus, M.A, Houdusse, A. | | Deposit date: | 2017-01-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Myosin 7 and its adaptors link cadherins to actin.
Nat Commun, 8, 2017
|
|
5MV8
 
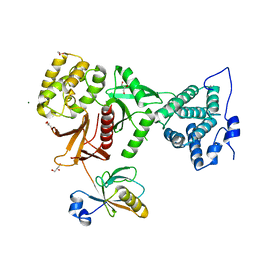 | | Structure of human Myosin 7b C-terminal MyTH4-FERM domain in complex with harmonin-a PDZ3 domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, Unconventional myosin-VIIb, ... | | Authors: | Yu, I, Sourigues, Y, Titus, M.A, Houdusse, A. | | Deposit date: | 2017-01-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Myosin 7 and its adaptors link cadherins to actin.
Nat Commun, 8, 2017
|
|
3AU5
 
 | |
4ZRK
 
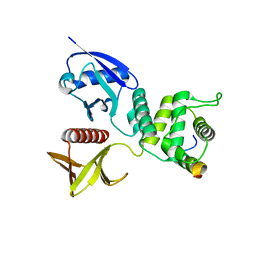 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4ZRI
 
 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4RM8
 
 | | Crystal structure of human ezrin in space group P21 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-20 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
3BIN
 
 | | Structure of the DAL-1 and TSLC1 (372-383) complex | | Descriptor: | Band 4.1-like protein 3, Cell adhesion molecule 1 | | Authors: | Busam, R.D, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Berglund, H, Persson, C, Hallberg, B.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B)
J.Biol.Chem., 286, 2011
|
|
4ZRJ
 
 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
8CIS
 
 | | The FERM domain of human moesin with two bound peptides identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C3P, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIU
 
 | | The FERM domain of human moesin mutant H288A | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIT
 
 | | The FERM domain of human moesin mutant L281R | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIR
 
 | | The FERM domain of human moesin with a bound peptide identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
4YL8
 
 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
4RM9
 
 | | Crystal structure of human ezrin in space group C2221 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
3WA0
 
 | | Crystal structure of merlin complexed with DCAF1/VprBP | | Descriptor: | Merlin, Protein VPRBP | | Authors: | Mori, T, Gotoh, S, Shirakawa, M, Hakoshima, T. | | Deposit date: | 2013-04-20 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of DDB1-and-Cullin 4-associated Factor 1 (DCAF1) recognition by merlin/NF2 and its implication in tumorigenesis by CD44-mediated inhibition of merlin suppression of DCAF1 function.
Genes Cells, 19, 2014
|
|
2ZPY
 
 | | Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide | | Descriptor: | CD44 antigen, Radixin | | Authors: | Mori, T, Kitano, K, Terawaki, S, Maesaki, R, Fukami, Y, Hakoshima, T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for CD44 recognition by ERM proteins
J.Biol.Chem., 283, 2008
|
|
3AU4
 
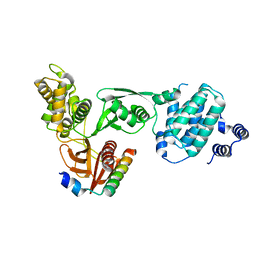 | |
4P7I
 
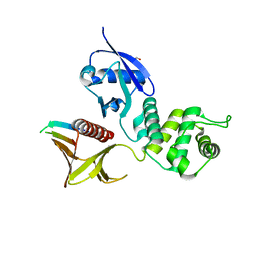 | | Crystal structure of the Merlin FERM/DCAF1 complex | | Descriptor: | GLYCEROL, Merlin, Protein VPRBP | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-03-27 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the binding of Merlin FERM domain to the E3 ubiquitin ligase substrate adaptor DCAF1.
J.Biol.Chem., 289, 2014
|
|
3X23
 
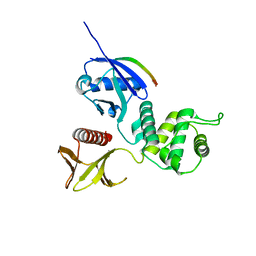 | | Radixin complex | | Descriptor: | Peptide from Matrix metalloproteinase-14, Radixin | | Authors: | Terawaki, S, Kitano, K, Aoyama, M, Mori, T, Hakoshima, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | MT1-MMP recognition by ERM proteins and its implication in CD44 shedding
Genes Cells, 20, 2015
|
|
4RMA
 
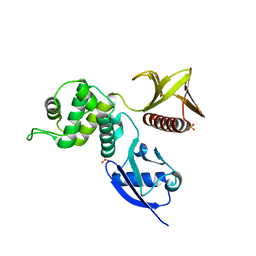 | | Crystal structure of the FERM domain of human ezrin | | Descriptor: | Ezrin, SULFATE ION | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
5XBF
 
 | | Crystal Structure of Myo7b C-terminal MyTH4-FERM in complex with USH1C PDZ3 | | Descriptor: | ACETATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Li, J, He, Y, Weck, W.L, Lu, Q, Tyska, M.J, Zhang, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
