4UYJ
 
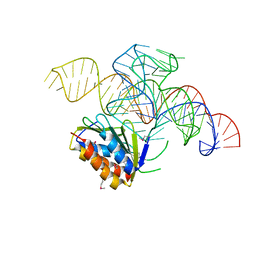 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
8C29
 
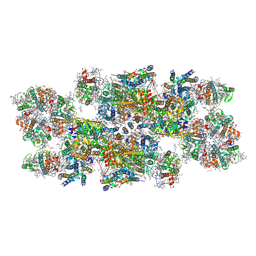 | | Cryo-EM structure of photosystem II C2S2 supercomplex from Norway spruce (Picea abies) at 2.8 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Kopecny, D, Semchonok, D.A, Kouril, R. | | Deposit date: | 2022-12-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.785 Å) | | Cite: | Cryo-EM structure of a plant photosystem II supercomplex with light-harvesting protein Lhcb8 and alpha-tocopherol.
Nat.Plants, 9, 2023
|
|
5WQH
 
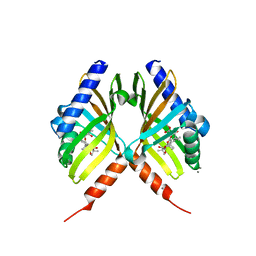 | | Structure of fungal meroterpenoid isomerase Trt14 complexed with substrate analog and endo-terretonin D | | Descriptor: | CALCIUM ION, Isomerase trt14, methyl (2S,4aR,4bS,5S,6aS,10aS,10bS,12aS)-2,4b,7,7,10a,12,12a-heptamethyl-5-oxidanyl-1,4,6,8-tetrakis(oxidanylidene)-4a,5,6a,9,10,10b-hexahydronaphtho[1,2-h]isochromene-2-carboxylate, ... | | Authors: | Mori, T, Matsuda, Y, Abe, I. | | Deposit date: | 2016-11-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis
Nat. Chem. Biol., 13, 2017
|
|
8IJN
 
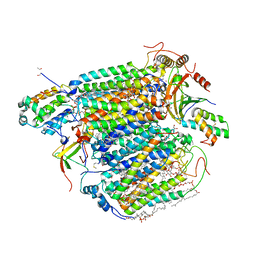 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A, Muramoto, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7TBT
 
 | |
6DHP
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.04 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
9BFY
 
 | | Tri-complex of Compound-12, KRAS G12C, and CypA | | Descriptor: | (3R)-N-[(2S)-1-{[(1M,8R,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methyl-1-propanoylpyrrolidine-3-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Tri-complex of Compound-12, KRAS G12C, and CypA
To be published
|
|
8CB0
 
 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 and NADP+ | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
8CAK
 
 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, DIMETHYL SULFOXIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
4MD2
 
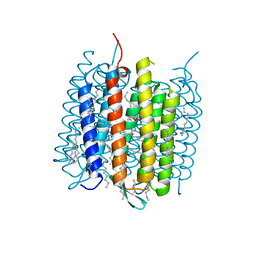 | | Ground state of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LTW
 
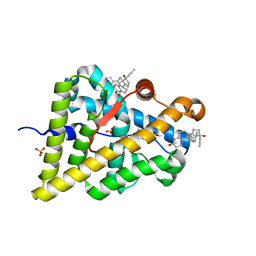 | | Ancestral Ketosteroid Receptor-Progesterone-Mifepristone Complex | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Ancestral Steroid Receptor 2, GLYCEROL, ... | | Authors: | Ortlund, E.A, Colucci, J.K. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | X-ray crystal structure of the ancestral 3-ketosteroid receptor-progesterone-mifepristone complex shows mifepristone bound at the coactivator binding interface.
Plos One, 8, 2013
|
|
4M2S
 
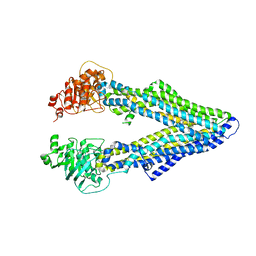 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-RRR | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
5EG4
 
 | | BOVINE TRYPSIN IN COMPLEX WITH CYCLIC INHIBITOR | | Descriptor: | (13S,16R)-N-(4-carbamimidoylbenzyl)-16-((N-cyclohexylsulfamoyl)amino)-3,9,15-trioxo-2,10,14-triaza-6(1,4)-piperazina-1, 11(1,4)-dibenzenacycloheptadecaphane-13-carboxamide, ACETATE ION, ... | | Authors: | Knoerlein, A, Wagner, S, Heine, A, Steinmetzer, T, Klebe, G. | | Deposit date: | 2015-10-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Optimization of Cyclic Plasmin Inhibitors: From Benzamidines to Benzylamines.
J.Med.Chem., 59, 2016
|
|
3NKN
 
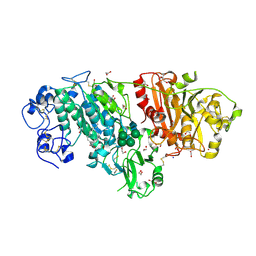 | | Crystal structure of mouse autotaxin in complex with 14:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
1JPO
 
 | | LOW TEMPERATURE ORTHORHOMBIC LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Bradbrook, G.M, Helliwell, J.R, Habash, J. | | Deposit date: | 1997-07-03 | | Release date: | 1997-11-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Time-Resolved Biological and Perturbation Chemical Crystallography: Laue and Monochromatic Developments
Time-Resolved Electron and X-Ray Diffraction; 13-14 July 1995, San Diego, California (in: Proc.Spie-Int.Soc.Opt.Eng., V.2521), 1995
|
|
1K4T
 
 | | HUMAN DNA TOPOISOMERASE I (70 KDA) IN COMPLEX WITH THE POISON TOPOTECAN AND COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Staker, B.L, Hjerrild, K, Feese, M.D, Behnke, C.A, Burgin Jr, A.B, Stewart, L.J. | | Deposit date: | 2001-10-08 | | Release date: | 2002-12-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of topoisomerase I poisoning by a camptothecin analog
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2W5J
 
 | | Structure of the c14-rotor ring of the proton translocating chloroplast ATP synthase | | Descriptor: | ATP SYNTHASE C CHAIN, CHLOROPLASTIC | | Authors: | Vollmar, M, Schlieper, D, Winn, M, Buechner, C, Groth, G. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase.
J. Biol. Chem., 284, 2009
|
|
1K43
 
 | | 10 Structure Ensemble of the 14-residue peptide RG-KWTY-NG-ITYE-GR (MBH12) | | Descriptor: | MBH12 | | Authors: | Pastor, M.T, Lopez de la Paz, M, Lacroix, E, Serrano, L, Perez-Paya, E. | | Deposit date: | 2001-10-05 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7O2M
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2289 | | Descriptor: | 1-[(3~{S},6~{S},9~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-9-(phenylmethyl)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, Genome polyprotein | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
2W8Y
 
 | | RU486 bound to the progesterone receptor in a destabilized agonistic conformation | | Descriptor: | (14beta,17alpha)-17-ethynyl-17-hydroxyestr-4-en-3-one, 1,2-ETHANEDIOL, 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, ... | | Authors: | Raaijmakers, H.C.A, Versteeg, J, Uitdehaag, J.C.M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Structure of Ru486 Bound to the Progesterone Receptor in a Destabilized Agonistic Conformation.
J.Biol.Chem., 284, 2009
|
|
4DPI
 
 | | BACE-1 in complex with HEA-macrocyclic inhibitor, MV078512 | | Descriptor: | (4S,8E,11R)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-16-methyl-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraene-2,13-dione, Beta-secretase 1 | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
1YJB
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 35% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YJK
 
 | | Reduced Peptidylglycine Alpha-Hydroxylating Monooxygenase (PHM) in a New Crystal Form | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Siebert, X, Eipper, B.A, Mains, R.E, Prigge, S.T, Blackburn, N.J, Amzel, L.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Catalytic Copper of Peptidylglycine alpha-Hydroxylating Monooxygenase also Plays a Critical Structural Role.
Biophys.J., 89, 2005
|
|
8Y37
 
 | | Cryo-EM structure of Staphylococcus aureus (15B196) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
8Y39
 
 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 70S ribosome in complex with MCX-190. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
