9C34
 
 | | Proline utilization A with the FADH- N5 atom covalently modified by proline | | Descriptor: | (2S)-3,4-dihydro-2H-pyrrole-2-carboxylic acid, (2~{S},5~{R})-5-[10-[(2~{S},3~{S},4~{R})-5-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-2,3,4-tris(oxidanyl)pentyl]-7,8-dimethyl-2,4-bis(oxidanylidene)benzo[g]pteridin-5-yl]pyrrolidine-2-carboxylic acid, Bifunctional protein PutA, ... | | Authors: | Tanner, J.J, Buckley, D.P. | | Deposit date: | 2024-05-31 | | Release date: | 2025-06-04 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Visualization of covalent intermediates and conformational states of proline utilization A by X-ray crystallography and molecular dynamics simulations.
J.Biol.Chem., 301, 2025
|
|
9C35
 
 | |
7T36
 
 | |
6SH6
 
 | | Crystal structure of the human DEAH-helicase DHX15 in complex with the NKRF G-patch bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jonas, S, Studer, M.K, Ivanovic, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for DEAH-helicase activation by G-patch proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SI0
 
 | | p53 cancer mutant Y220C in complex with small-molecule stabilizer PK9323 | | Descriptor: | 1,2-ETHANEDIOL, 1-[9-ethyl-7-(1,3-thiazol-4-yl)carbazol-3-yl]-~{N}-methyl-methanamine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Joerger, A.C, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
8FI4
 
 | |
6HVY
 
 | | Yeast 20S proteasome in complex with 5 (7- and 6-membered ring) | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[(1~{S},4~{a}~{S},8~{a}~{R})-1,2,3,4,4~{a},5,6,7,8,8~{a}-decahydronaphthalen-1-yl]-4-methyl-3,4-bis(oxidanyl)pentan-2-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, (2~{S})-~{N}-[(2~{S},3~{S},4~{R})-1-[(1~{S},4~{a}~{S},8~{a}~{R})-1,2,3,4,4~{a},5,6,7,8,8~{a}-decahydronaphthalen-1-yl]-4-methyl-3,5-bis(oxidanyl)pentan-2-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
8C54
 
 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
6HW7
 
 | | Yeast 20S proteasome in complex with 29 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
8JU9
 
 | |
5NJ5
 
 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
8THH
 
 | | Cryo-EM structure of Nav1.7 with LTG | | Descriptor: | (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9GIO
 
 | |
7ZVB
 
 | | Crystal Structure of the mature form of the glutamic-class prolyl-endopeptidase neprosin at 2.35 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptidase, TRIETHYLENE GLYCOL, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
9DVO
 
 | | Structure of the phosphate exporter XPR1/SLC53A1, Pi and InsP8-bound, intracellular gate open/intracellular gate closed state | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
9DVN
 
 | | Structure of the phosphate exporter XPR1/SLC53A1, InsP8-bound, intracellular gate closed state | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
8Z82
 
 | | Photosynthetic LH1-RC-HiPIP complex from the purple bacterium Halorhodospira halophila | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna complex, alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Nagashima, K.V.P, Hiwatashi, N, Kawakami, M, Nakata, K, Nagashima, S, Inoue, K, Takaichi, S, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-29 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A Native LH1-RC-HiPIP Supercomplex from an Extremophilic Phototroph.
Commun Biol, 8, 2025
|
|
8K8J
 
 | | Cannabinoid Receptor 1 bound to Fenofibrate coupling MiniGsq and Nb35 Complex | | Descriptor: | Fenofibrate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tang, W.Q, Wang, T.X, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Fenofibrate Recognition and G q Protein Coupling Mechanisms of the Human Cannabinoid Receptor CB1.
Adv Sci, 11, 2024
|
|
7S0S
 
 | | M. tuberculosis ribosomal RNA methyltransferase TlyA bound to M. smegmatis 50S ribosomal subunit | | Descriptor: | 16S/23S rRNA (Cytidine-2'-O)-methyltransferase TlyA, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Laughlin, Z.T, Dunham, C.M, Conn, G.L. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | 50S subunit recognition and modification by the Mycobacterium tuberculosis ribosomal RNA methyltransferase TlyA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9DVM
 
 | | Structure of the phosphate exporter XPR1/SLC53A1, InsP8-bound, intracellular gate open/intracellular gate closed state | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
5K2Z
 
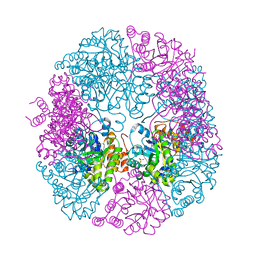 | | PDX1.3-adduct (Arabidopsis) | | Descriptor: | 1,2-ETHANEDIOL, 2-azanylpenta-1,4-dien-3-one, CHLORIDE ION, ... | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4OC0
 
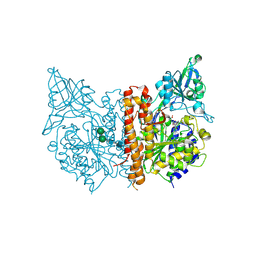 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CCIBzL, a urea-based inhibitor N~2~-[(1-carboxycyclopropyl)carbamoyl]-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5NFH
 
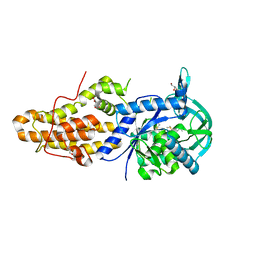 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with a quinazolinone inhibitor | | Descriptor: | 2-[3-[[4,6-bis(chloranyl)-1~{H}-indol-2-yl]methylamino]propylamino]-3~{H}-quinazolin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Robinson, D.A, Eadsforth, T.C, Shepherd, S.M, Torrie, L.S, De Rycker, M, Gilbert, I.H. | | Deposit date: | 2017-03-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemical Validation of Methionyl-tRNA Synthetase as a Druggable Target in Leishmania donovani.
ACS Infect Dis, 3, 2017
|
|
6GNF
 
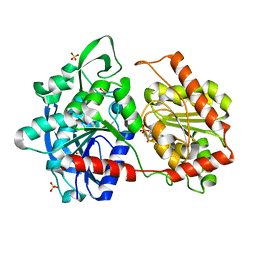 | | Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase, ... | | Authors: | Cuesta-Seijo, J.A, Nielsen, M.M, Palcic, M.M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of theCatalyticDomain ofArabidopsis thalianaStarch Synthase IV, of Granule Bound Starch Synthase From CLg1 and of Granule Bound Starch Synthase I ofCyanophora paradoxaIllustrate Substrate Recognition in Starch Synthases.
Front Plant Sci, 9, 2018
|
|
8JXR
 
 | | Structure of nanobody-bound DRD1_LSD complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 112, 2024
|
|
