6YH2
 
 | | tRNA-guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Co-crystallization, nanoESI-MS and 19F NMR reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
7QL7
 
 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 9 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8VG9
 
 | |
8CPA
 
 | |
8QXZ
 
 | | Xylanase from Bacillus circulans mutant E78Q/Y69A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX.
Febs J., 291, 2024
|
|
4ILV
 
 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
8QY2
 
 | | Xylanase from Bacillus circulans mutant E78Q/F125A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Chikunova, A, Saberi, M, Ubbink, M. | | Deposit date: | 2023-10-25 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bimodal substrate binding in the active site of the glycosidase BcX.
Febs J., 291, 2024
|
|
5J6L
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with N-Butyl-5-[4-(2-fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | Heat shock protein HSP 90-alpha, N-butyl-5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methylbenzamide | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
8V5H
 
 | |
5ZNK
 
 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-chloro-6-fluoro-3-{2-oxo-3-[(2S)-piperidin-2-yl]propyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
8VFQ
 
 | | De novo design apixaban-binding protein: apx1049 | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, De novo designed apixaban-binding protein apx1049 | | Authors: | Bera, A.K, Lee, G.R, Baker, D. | | Deposit date: | 2023-12-21 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-molecule binding and sensing with a designed protein family
To Be Published
|
|
8C6V
 
 | |
8CG7
 
 | | Structure of p53 cancer mutant Y220C with arylation at Cys182 and Cys277 | | Descriptor: | 1,2-ETHANEDIOL, 5-iodanyl-2-methylsulfonyl-pyrimidine, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Pichon, M.M, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Reactivity Studies of 2-Sulfonylpyrimidines Allow Selective Protein Arylation.
Bioconjug.Chem., 34, 2023
|
|
8R2B
 
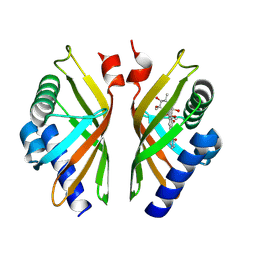 | |
8HOZ
 
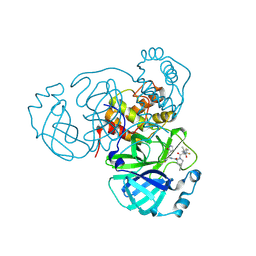 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
7T5J
 
 | | Crystal Structure of Strictosidine Synthase in complex with S-IPA (2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2S)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
7SZC
 
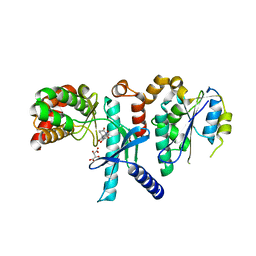 | |
7SZD
 
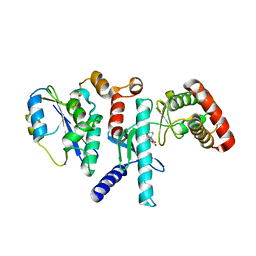 | |
6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
7T5I
 
 | | Crystal Structure of Strictosidine Synthase in complex with R-IPA(2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2R)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
5CIP
 
 | | Crystal Structure of Unbound 4E10 | | Descriptor: | FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
8A1N
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with fumaryl amide analogue 13 | | Descriptor: | (Z)-N-(4-chlorophenyl)-4-oxidanylidene-but-2-enamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
6G36
 
 | | Crystal structure of haspin in complex with 5-chlorotubercidin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chlorotubercidin, COBALT (II) ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8TMA
 
 | | Antibody N3-1 bound to RBD in the up conformation | | Descriptor: | N3-1 Fab heavy chain, N3-1 Fab light chain, Spike glycoprotein | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SARS-COV-2 Omicron variants conformationally escape a rare quaternary antibody binding mode.
Commun Biol, 6, 2023
|
|
