2O9J
 
 | | Crystal structure of calcium atpase with bound magnesium fluoride and cyclopiazonic acid | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Young, H.S, Moncoq, K.A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The molecular basis for cyclopiazonic Acid inhibition of the sarcoplasmic reticulum calcium pump.
J.Biol.Chem., 282, 2007
|
|
2O9K
 
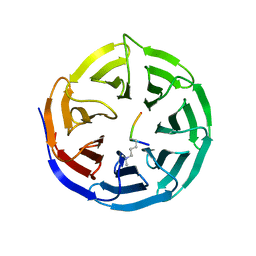 | | WDR5 in Complex with Dimethylated H3K4 Peptide | | Descriptor: | H3 HISTONE, WD repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Molecular Recognition and Presentation of Histone H3 by Wdr5.
Embo J., 25, 2006
|
|
2O9L
 
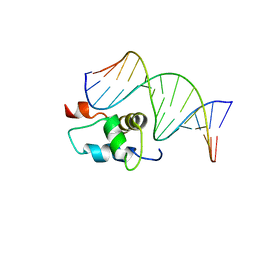 | | AMBER refined NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | | Descriptor: | 5'-D(*GP*AP*AP*AP*CP*GP*TP*GP*CP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*GP*CP*AP*CP*GP*TP*TP*TP*C)-3', RNA polymerase sigma factor RpoN | | Authors: | Doucleff, M, Pelton, J.G, Lee, P.S, Wemmer, D.E. | | Deposit date: | 2006-12-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA recognition by the alternative sigma-factor, sigma54.
J.Mol.Biol., 369, 2007
|
|
2O9O
 
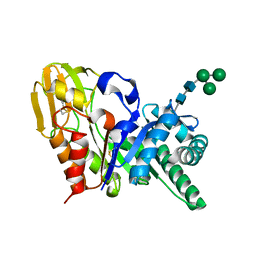 | | Crystal Structure of the buffalo Secretory Signalling Glycoprotein at 2.8 A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ethayathulla, A.S, Srivastava, D.B, Kumar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the buffalo secretory signalling glycoprotein at 2.8 A resolution
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2O9P
 
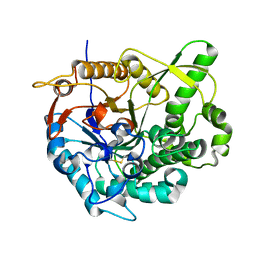 | | beta-glucosidase B from Paenibacillus polymyxa | | Descriptor: | Beta-glucosidase B | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
2O9Q
 
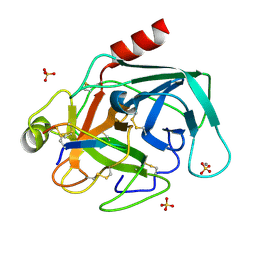 | | The crystal structure of Bovine Trypsin complexed with a small inhibition peptide ORB2K | | Descriptor: | CALCIUM ION, Cationic trypsin, ORB2K, ... | | Authors: | Li, J, Zhang, C, Xu, X, Wang, J, Gong, W, Lai, R. | | Deposit date: | 2006-12-14 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From protease inhibitor to antibiotics: single point mutation makes tremendous functional shift
To be Published
|
|
2O9R
 
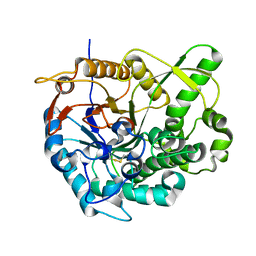 | | beta-glucosidase B complexed with thiocellobiose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
2O9S
 
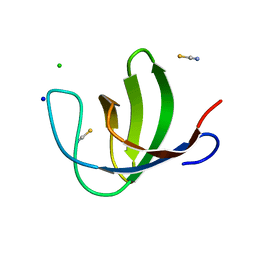 | | The second SH3 domain from ponsin | | Descriptor: | CHLORIDE ION, Ponsin, SODIUM ION, ... | | Authors: | Pinotsis, N, Wilmanns, M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Paxillin and ponsin interact in nascent costameres of muscle cells
J.Mol.Biol., 369, 2007
|
|
2O9T
 
 | |
2O9U
 
 | |
2O9V
 
 | |
2O9X
 
 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | Descriptor: | Reductase, assembly protein | | Authors: | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2O9Z
 
 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the Apo form | | Descriptor: | PHOSPHATE ION, Tryptophan halogenase | | Authors: | Bitto, E, Bingman, C.A, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
2OA0
 
 | | Crystal structure of Calcium ATPase with bound ADP and cyclopiazonic acid | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Young, H.S, Moncoq, K.A. | | Deposit date: | 2006-12-14 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The molecular basis for cyclopiazonic Acid inhibition of the sarcoplasmic reticulum calcium pump.
J.Biol.Chem., 282, 2007
|
|
2OA1
 
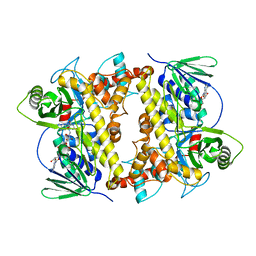 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the L-Tryptophan with FAD complex | | Descriptor: | ADENOSINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bitto, E, Bingman, C.A, Singh, S, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
2OA2
 
 | |
2OA4
 
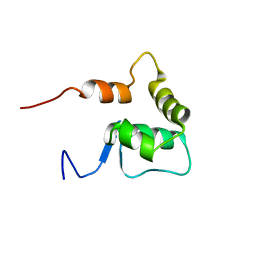 | | Solution NMR Structure: Northeast Structural Genomics Consortium Target SiR5 | | Descriptor: | SiR5 | | Authors: | Wang, L, Rossi, P, Chen, C.X, Nwosu, C, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Burkhard, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target SiR5
To be Published
|
|
2OA5
 
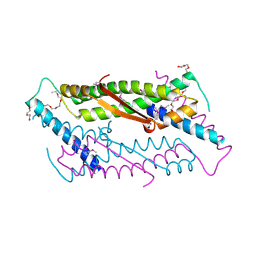 | | Crystal structure of ORF52 from Murid herpesvirus (MUHV-4) (Murine gammaherpesvirus 68) at 2.1 A resolution. Northeast Structural Genomics Consortium target MHR28B. | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Hypothetical protein BQLF2 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Ho, C.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68.
J.Biol.Chem., 282, 2007
|
|
2OA6
 
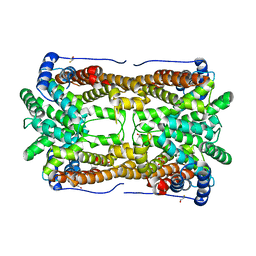 | | Aristolochene synthase from Aspergillus terreus complexed with pyrophosphate | | Descriptor: | Aristolochene synthase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Shishova, E.Y, Di Costanzo, L, Cane, D.E, Christianson, D.W. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate
Biochemistry, 46, 2007
|
|
2OA7
 
 | |
2OA8
 
 | | Crystal Structure of mTREX1 with ssDNA | | Descriptor: | 5'-D(*GP*AP*CP*G)-3', CALCIUM ION, Three prime repair exonuclease 1 | | Authors: | de Silva, U, Hollis, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of TREX1 Explains the 3' Nucleotide Specificity and Reveals a Polyproline II Helix for Protein Partnering.
J.Biol.Chem., 282, 2007
|
|
2OA9
 
 | | Restriction endonuclease MvaI in the absence of DNA | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kaus-Drobek, M, Czapinska, H, Sokolowska, M, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically.
Nucleic Acids Res., 35, 2007
|
|
2OAA
 
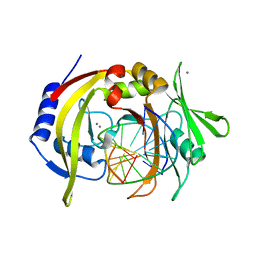 | | Restriction endonuclease MvaI-cognate DNA substrate complex | | Descriptor: | 5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3', CALCIUM ION, ... | | Authors: | Kaus-Drobek, M, Czapinska, H, Sokolowska, M, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically.
Nucleic Acids Res., 35, 2007
|
|
2OAC
 
 | |
2OAD
 
 | |
