2G7F
 
 | | The 1.95 A crystal structure of Vibrio cholerae extracellular endonuclease I | | Descriptor: | CHLORIDE ION, Endonuclease I, MAGNESIUM ION | | Authors: | Altermark, B, Smalaas, A.O, Willassen, N.P, Helland, R. | | Deposit date: | 2006-02-28 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Vibrio cholerae extracellular endonuclease I reveals the presence of a buried chloride ion.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2F98
 
 | | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. | | Descriptor: | Aklanonic Acid methyl Ester Cyclase, AknH, METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, ... | | Authors: | Kallio, P, Sultana, A, Neimi, J, Mantsala, P, Schneider, G. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity.
J.Mol.Biol., 357, 2006
|
|
2H02
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Protein tyrosine phosphatase, receptor type, B,, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HC1
 
 | | Engineered catalytic domain of protein tyrosine phosphatase HPTPbeta. | | Descriptor: | ACETATE ION, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2H04
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Protein tyrosine phosphatase, receptor type, B,, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HC2
 
 | | Engineered protein tyrosine phosphatase beta catalytic domain | | Descriptor: | MAGNESIUM ION, Receptor-type tyrosine-protein phosphatase beta, SODIUM ION | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2VFV
 
 | | Alditol Oxidase from Streptomyces coelicolor A3(2): Complex with Sulphite | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, CHLORIDE ION, XYLITOL OXIDASE | | Authors: | Forneris, F, Mattevi, A. | | Deposit date: | 2007-11-05 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Stereoselectivity in Streptomyces Coelicolor Alditol Oxidase.
Biochemistry, 47, 2008
|
|
3E2D
 
 | | The 1.4 A crystal structure of the large and cold-active Vibrio sp. alkaline phosphatase | | Descriptor: | 1,2-ETHANEDIOL, Alkaline phosphatase, MAGNESIUM ION, ... | | Authors: | Helland, R, Larsen, R.L, Asgeirsson, B. | | Deposit date: | 2008-08-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4 A crystal structure of the large and cold-active Vibrio sp. alkaline phosphatase.
Biochim.Biophys.Acta, 1794, 2009
|
|
3IN1
 
 | |
3IH0
 
 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized sugar kinase PH1459 | | Authors: | Kumar, G, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-29 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP
To be Published
|
|
3EHA
 
 | |
3IKH
 
 | |
2XAW
 
 | |
3IE7
 
 | |
2VND
 
 | | The N69Q mutant of Vibrio cholerae endonuclease I | | Descriptor: | CHLORIDE ION, ENDONUCLEASE I, MAGNESIUM ION | | Authors: | Niiranen, L, Altermark, B, Brandsdal, B.O, Leiros, H.-K.S, Helland, R, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effects of Salt on the Kinetics and Thermodynamic Stability of Endonuclease I from Vibrio Salmonicida and Vibrio Cholerae.
FEBS J., 275, 2008
|
|
3EWD
 
 | | Crystal structure of adenosine deaminase mutant (delta Asp172) from Plasmodium vivax in complex with MT-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, Adenosine deaminase, ZINC ION | | Authors: | Schramm, V.L, Almo, S.C, Cassera, M.B, Ho, M.C. | | Deposit date: | 2008-10-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
Biochemistry, 48, 2009
|
|
3ILH
 
 | |
2XAV
 
 | |
2XAX
 
 | |
2X0X
 
 | | Ribonucleotide reductase R1 subunit of E. coli to 2.3 A resolution | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA, SULFATE ION | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-Specific Incorporation of 3-Nitrotyrosine as a Probe of Pk(A) Perturbation of Redox-Active Tyrosines in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
3KD2
 
 | | Crystal structure of the CFTR inhibitory factor Cif | | Descriptor: | CFTR inhibitory factor (Cif) | | Authors: | Bahl, C.D, Madden, D.R. | | Deposit date: | 2009-10-22 | | Release date: | 2010-01-26 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor.
J.Bacteriol., 192, 2010
|
|
3F5G
 
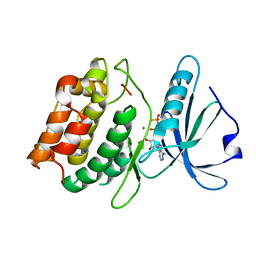 | | Crystal structure of death associated protein kinase in complex with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, MAGNESIUM ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-03 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3KMN
 
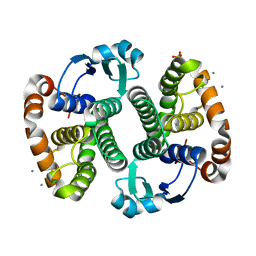 | | Crystal Structure of the Human Apo GST Pi C47S/Y108V Double Mutant | | Descriptor: | CALCIUM ION, CARBONATE ION, Glutathione S-transferase P, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diuretic drug binding to human glutathione transferase P1-1: potential role of CYS101 revealed in the double mutant C47S/Y108V
to be published
|
|
3F5U
 
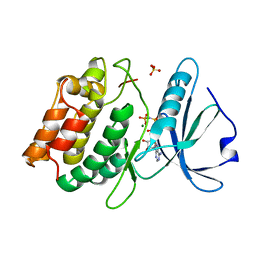 | | Crystal structure of the death associated protein kinase in complex with AMPPNP and Mg2+ | | Descriptor: | Death-associated protein kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3KM6
 
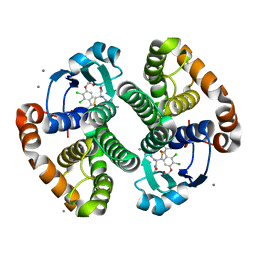 | |
