7ZK1
 
 | |
7R0W
 
 | | 2.8 Angstrom cryo-EM structure of the dimeric cytochrome b6f-PetP complex from Synechocystis sp. PCC 6803 with natively bound lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S,8E)-1-{[(2S)-1-hydroxy-3-{[(1S)-1-hydroxypentadecyl]oxy}propan-2-yl]oxy}heptadec-8-en-1-ol, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, ... | | Authors: | Farmer, D.F, Proctor, M.S, Malone, L.A, Swainsbury, D.P.K, Hawkings, F.R, Hitchcock, A, Johnson, M.P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the Synechocystis sp. PCC 6803 cytochrome b6f complex with and without the regulatory PetP subunit.
Biochem.J., 479, 2022
|
|
6RZ8
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6RZ9
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2770372 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(5-fluoranyl-2-methyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3-dih ydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6RZ6
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (C2221 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
4LVC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, AMMONIUM ION, ... | | Authors: | Manszewski, T, Singh, K, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An enzyme captured in two conformational states: crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1GD4
 
 | | SOLUTION STRUCTURE OF P25S CYSTATIN A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
1GD3
 
 | | refined solution structure of human cystatin A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix
conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
5YFP
 
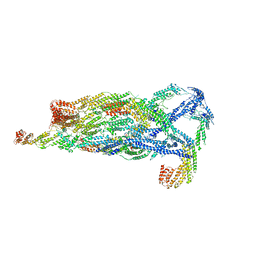 | | Cryo-EM Structure of the Exocyst Complex | | Descriptor: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | Authors: | Mei, K, Li, Y, Wang, S, Shao, G, Wang, J, Ding, Y, Luo, G, Yue, P, Liu, J.J, Wang, X, Dong, M.Q, Guo, W, Wang, H.W. | | Deposit date: | 2017-09-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the exocyst complex
Nat. Struct. Mol. Biol., 25, 2018
|
|
3TX3
 
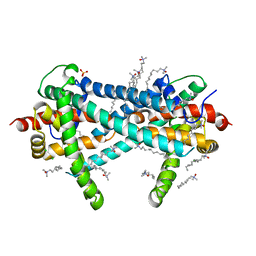 | | CysZ, a putative sulfate permease | | Descriptor: | CHLORIDE ION, LAURYL DIMETHYLAMINE-N-OXIDE, SULFATE ION, ... | | Authors: | Assur, Z, Liu, Q, Hendrickson, W.A, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CysZ, a putative sulfate permease
To be Published
|
|
2B1E
 
 | | The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif | | Descriptor: | Exocyst complex component EXO70 | | Authors: | Dong, G, Hutagalung, A.H, Fu, C, Novick, P, Reinisch, K.M. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif
Nat.Struct.Mol.Biol., 12, 2005
|
|
6WUE
 
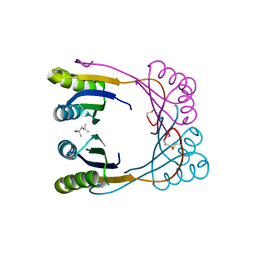 | | Tetragonal crystal form of SbtB from Synechocystis PCC6803 | | Descriptor: | ISOPROPYL ALCOHOL, Membrane-associated protein slr1513, SODIUM ION | | Authors: | Bu, G, Simmons, C.R, Nielsen, D.R, Nannenga, B.L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tetragonal crystal form of the cyanobacterial bicarbonate-transporter regulator SbtB from Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6H0C
 
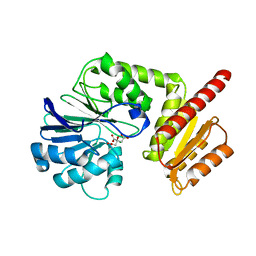 | | Flv1 flavodiiron core from Synechocystis sp. PCC6803 | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative diflavin flavoprotein A 3 | | Authors: | Borges, P.T, Romao, C.V, Saraiva, L, Goncalves, V.L, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2018-07-08 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Analysis of a new flavodiiron core structural arrangement in Flv1-Delta FlR protein from Synechocystis sp. PCC6803.
J. Struct. Biol., 205, 2019
|
|
6H0D
 
 | | Metal soaked Flv1 flavodiiron core from Synechocystis sp. PCC6803 | | Descriptor: | CHLORIDE ION, Putative diflavin flavoprotein A 3, SULFATE ION | | Authors: | Borges, P.T, Romao, C.V, Saraiva, L, Goncalves, V.L, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2018-07-08 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Analysis of a new flavodiiron core structural arrangement in Flv1-Delta FlR protein from Synechocystis sp. PCC6803.
J. Struct. Biol., 205, 2019
|
|
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6T9O
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
5DJK
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PO4 and 2Ca bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
2ZJ0
 
 | | Crystal structure of Mycobacterium tuberculosis S-Adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
5DMM
 
 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Metallated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, BETA-MERCAPTOETHANOL, Homocysteine S-methyltransferase, ... | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
8WZ7
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(I255A,T287A) in ternary complex with NAD and adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WZ9
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and adenine | | Descriptor: | ADENINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
2ZIZ
 
 | | Crystal structure of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 3-deazaadenosine | | Descriptor: | 3-DEAZA-ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
6BVE
 
 | |
5EPE
 
 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
5DML
 
 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Oxidized form | | Descriptor: | CHLORIDE ION, Homocysteine S-methyltransferase | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
