8ZPP
 
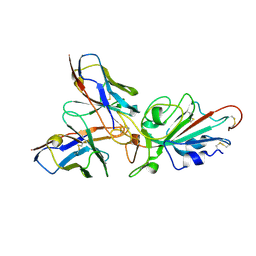 | | Local CryoEM structure of the SARS-CoV-2 BA.5 in complex with ORB10 Fab | | Descriptor: | Spike glycoprotein,Fibritin, variable heavy chain of ORB10 Fab, variable light chain of ORB10 Fab | | Authors: | Cao, S, Leng, C, Hu, H. | | Deposit date: | 2024-05-30 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into hybridoma-derived neutralizing monoclonal antibodies against Omicron BA.5 and XBB.1.16 variants of SARS-CoV-2.
J.Virol., 99, 2025
|
|
9KTO
 
 | |
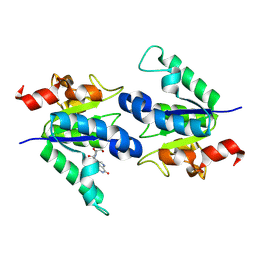
8PQP
 
 | |
7PJG
 
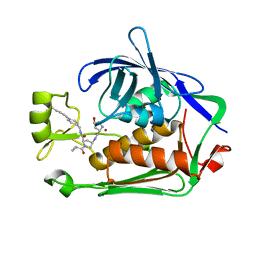 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2R,4R)-N-[[4-(4-cyclopropylbuta-1,3-diynyl)phenyl]methyl]-1-(2-methylpropanoyl)-4-[[(2S,4R)-4-oxidanylpyrrolidin-2-yl]carbonylamino]pyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
9KTM
 
 | |
8U7W
 
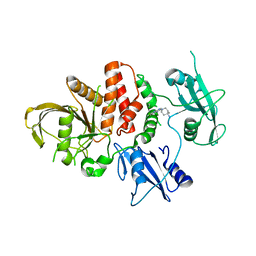 | |
6T2O
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16 | | Authors: | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | Deposit date: | 2019-10-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
7PK8
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2S,4S)-4-fluoranyl-N-[(3R,5R)-5-[[[4-[2-(4-methylphenyl)ethynyl]phenyl]carbonylamino]methyl]-1-(2-methylpropanoyl)pyrrolidin-3-yl]pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
8GX4
 
 | |
8UFS
 
 | | Structure of human endothelial nitric oxide synthase E361Q mutant heme domain obtain after soaking crystal with 4-methyl-7-(4-methyl-2,3,4,5-tetrahydrobenzo[f][1,4]oxazepin-7-yl)quinolin-2-amine dihydrochloride | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, CALCIUM ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
6QCN
 
 | | Human Sirt2 in complex with ADP-ribose and the inhibitor quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Riemer, S, You, W, Steegborn, C. | | Deposit date: | 2018-12-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives.
Sci Rep, 9, 2019
|
|
6HOV
 
 | |
6YJH
 
 | | Crystal structure of Imidazole Glycerol Phosphate Dehydratase from Mycobacterium tuberculosis at 1.61 A resolution | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Timofeev, V.I, Abramchik, Y.A, Skvortsov, T.A, Azhikina, T.L, Muravieva, T.I, Kostromina, M.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of Imidazole Glycerol Phosphate Dehydratase from Mycobacterium tuberculosis at 1.61 A resolution
To Be Published
|
|
6NCL
 
 | |
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
8UIW
 
 | | yjdF riboswitch from R. gauvreauii in complex with chelerythrine bound to Fab BL3-6 S97N | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, ... | | Authors: | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | Deposit date: | 2023-10-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
8XLD
 
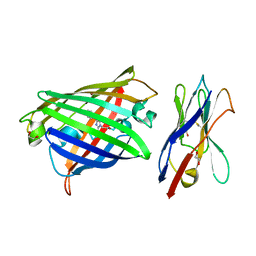 | | Structure of the GFP:GFP-nanobody complex from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Nanobody(Staygold-S2G10)-Nanobody(Staygold-S4F1), ZINC ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the GFP:GFP-nanobody complex from Biortus.
To Be Published
|
|
8BFS
 
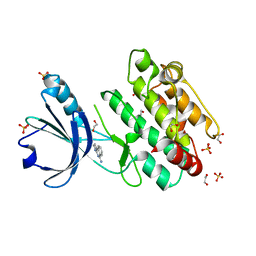 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326 | | Descriptor: | 1,2-ETHANEDIOL, 3~{H}-pyrrolo[2,3-c]isoquinolin-5-amine, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Zhu, W.F, Hernandez-Olmos, V, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326
To Be Published
|
|
9LTK
 
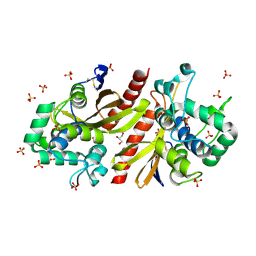 | | Crystal structur of Lpg1618(R215F) from Legionella pneumophila | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Liu, T, Gao, J, Ge, H. | | Deposit date: | 2025-02-06 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular characterization of AmpS, a class D beta-lactamase from Legionella pneumophila.
Int.J.Biol.Macromol., 312, 2025
|
|
8UOO
 
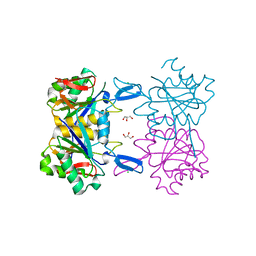 | | Structure of atypical asparaginase from Rhodospirillum rubrum | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
6NPV
 
 | |
6WCD
 
 | | Crystal Structure of Xenopus laevis APE2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Endogenous DNA 3' Blocks Are Vulnerabilities for BRCA1 and BRCA2 Deficiency and Are Reversed by the APE2 Nuclease.
Mol.Cell, 78, 2020
|
|
6I1F
 
 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Penicillin-binding protein,Penicillin-binding protein | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin
To Be Published
|
|
7PSF
 
 | |
7AM1
 
 | | Structure of yeast Ssd1, a pseudonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protein SSD1 | | Authors: | Cook, A.G, Jayachandran, U. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast Ssd1 is a non-enzymatic member of the RNase II family with an alternative RNA recognition site.
Nucleic Acids Res., 50, 2022
|
|
