9MQC
 
 | | Vitamin K-dependent gamma-carboxylase with Osteocalcin (mutant) and vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2025-01-02 | | Release date: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into the vitamin K-dependent gamma-carboxylation of osteocalcin.
Cell Res., 35, 2025
|
|
2L1J
 
 | | 1H assignments for ASIP(93-126, P103A, P105A, P111A, Q115Y, S124Y) | | Descriptor: | Agouti-signaling protein | | Authors: | Patel, M.P, Cribb Fabersunne, C.S, Yang, Y, Kaelin, C.B, Barsh, G.S, Millhauser, G.L. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Loop-swapped chimeras of the agouti-related protein and the agouti signaling protein identify contacts required for melanocortin 1 receptor selectivity and antagonism.
J.Mol.Biol., 404, 2010
|
|
9NMQ
 
 | | Structure of mouse RyR1 with simvastatin (Ca2+/CFF/ATP dataset; closed pore) | | Descriptor: | (1S,3R,7S,8S,8aR)-8-{2-[(2R,4R)-4-hydroxy-6-oxooxan-2-yl]ethyl}-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl 2,2-dimethylbutanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2025-03-04 | | Release date: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for statin-induced skeletal muscle weakness
To Be Published
|
|
7L08
 
 | | Cryo-EM structure of the human 55S mitoribosome-RRFmt complex. | | Descriptor: | 12S rRNA, 16S RRNA, 28S ribosomal protein S10, ... | | Authors: | Koripella, R, Agrawal, E.K, Deep, A, Agrawal, R.K. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Distinct mechanisms of the human mitoribosome recycling and antibiotic resistance.
Nat Commun, 12, 2021
|
|
7VEK
 
 | | Crystal structure of Phytolacca americana UGT3 with capsaicin and UDP-2fluoroglucose | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4GTM
 
 | | FTase in complex with BMS analogue 11 | | Descriptor: | 4-({(3R)-7-cyano-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl hexylcarbamate, DIMETHYL SULFOXIDE, FARNESYL DIPHOSPHATE, ... | | Authors: | Guo, Z, Stigter, E.A, Bon, R.S, Waldmann, H, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Selective, Potent RabGGTase Inhibitors
J.Med.Chem., 55, 2012
|
|
6UVY
 
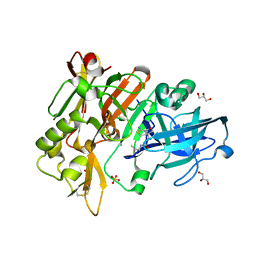 | | BACE-1 in complex with compound #18 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6I2S
 
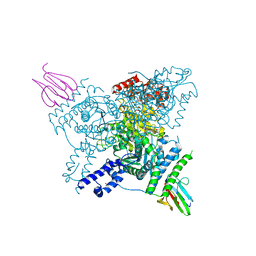 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD (R802A) in complex with GarA, following 2-oxoglutarate soak | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, CALCIUM ION, Glycogen accumulation regulator GarA, ... | | Authors: | Wagner, T, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the functional versatility of an FHA domain protein in mycobacterial signaling.
Sci.Signal., 12, 2019
|
|
6I76
 
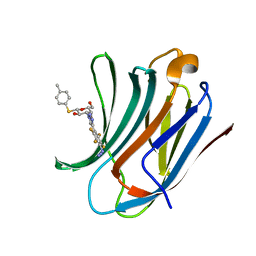 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-3 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azido-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6V1B
 
 | | Crystal structure of the bromodomain of human BRD9 bound to I-BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
3BJW
 
 | | Crystal Structure of ecarpholin S complexed with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Phospholipase A2 | | Authors: | Zhou, X, Sivaraman, J. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
6IBE
 
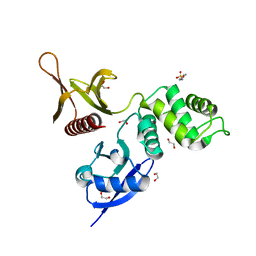 | | The FERM domain of Human EPB41L3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Band 4.1-like protein 3 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The FERM domain of Human EPB41L3
To Be Published
|
|
4ZEA
 
 | | Endothiapepsin in complex with fragment B91 | | Descriptor: | 1,2-ETHANEDIOL, 2-IMINOBIOTIN, ACETATE ION, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-04-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7VDQ
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 7 | | Descriptor: | 2-[[7-[[2-fluoranyl-4-[3-(hydroxymethyl)pyrazol-1-yl]phenyl]amino]-1,6-naphthyridin-2-yl]-(1-methylpiperidin-4-yl)amino]ethanoic acid, Cyclin-dependent kinase 5 activator 1, p25, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VTQ
 
 | | Cryo-EM structure of mouse NLRP3 (full-length) dodecamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2021-10-30 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4ZFV
 
 | | Lipomyces starkeyi levoglucosan kinase bound to ADP and magnesium. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Levoglucosan kinase, ... | | Authors: | Bacik, J.P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Producing Glucose 6-Phosphate from Cellulosic Biomass: STRUCTURAL INSIGHTS INTO LEVOGLUCOSAN BIOCONVERSION.
J.Biol.Chem., 290, 2015
|
|
8XSD
 
 | | BA.5 Spike complex with CR9 | | Descriptor: | CR9 heavy chain, CR9 light chain, Spike glycoprotein | | Authors: | Feng, L.L. | | Deposit date: | 2024-01-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | A broadly neutralizing antibody against the SARS-CoV-2 Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.4, and BA.5.
Signal Transduct Target Ther, 10, 2025
|
|
6IH0
 
 | | Aquifex aeolicus LpxC complex with ACHN-975 | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Li, D.Y, Fan, S, Jin, Y.Y, Zhang, C, Lv, G.X, Wu, G.T, Yang, Z.Y. | | Deposit date: | 2018-09-27 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Complex Structure of Protein AaLpxC from Aquifex aeolicus with ACHN-975 Molecule Suggests an Inhibitory Mechanism at Atomic-Level against Gram-Negative Bacteria
Molecules, 26, 2021
|
|
8X48
 
 | | Cryo-EM structure of Ryanodine receptor 1 (EGTA) | | Descriptor: | Ryanodine receptor 1 | | Authors: | Chen, Q, Hu, H. | | Deposit date: | 2023-11-15 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into transmembrane helix S0 facilitated RyR1 channel gating by Ca 2+ /ATP.
Nat Commun, 16, 2025
|
|
6UVV
 
 | | BACE-1 in complex with compound #17 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-butylcyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
8X49
 
 | | Cryo-EM structure of Ryanodine receptor 1 (100 nM Ca2+) | | Descriptor: | CALCIUM ION, Ryanodine receptor 1, ZINC ION | | Authors: | Chen, Q, Hu, H. | | Deposit date: | 2023-11-15 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into transmembrane helix S0 facilitated RyR1 channel gating by Ca 2+ /ATP.
Nat Commun, 16, 2025
|
|
4ZCU
 
 | | Structure of calcium-bound regulatory domain of the human ATP-Mg/Pi carrier in the P2 form | | Descriptor: | CALCIUM ION, Calcium-binding mitochondrial carrier protein SCaMC-1, GLYCEROL, ... | | Authors: | Harborne, S.P.D, Ruprecht, J.J, Kunji, E.R.S. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Calcium-induced conformational changes in the regulatory domain of the human mitochondrial ATP-Mg/Pi carrier.
Biochim.Biophys.Acta, 1847, 2015
|
|
6V91
 
 | |
7Q3Y
 
 | | Structure of full-length, monomeric, soluble somatic angiotensin I-converting enzyme showing the N- and C-terminal ellipsoid domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
3C5W
 
 | | Complex between PP2A-specific methylesterase PME-1 and PP2A core enzyme | | Descriptor: | PP2A A subunit, PP2A C subunit, PP2A-specific methylesterase PME-1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
