2OEJ
 
 | | Crystal structure of a rubisco-like protein from Geobacillus kaustophilus (tetramutant form), liganded with phosphate ions | | Descriptor: | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase, PHOSPHATE ION | | Authors: | Fedorov, A.A, Imker, H.J, Fedorov, E.V, Almo, S.C, Gerlt, J.A. | | Deposit date: | 2006-12-30 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanistic Diversity in the RuBisCO Superfamily: The "Enolase" in the Methionine Salvage Pathway in Geobacillus kaustophilus.
Biochemistry, 46, 2007
|
|
2OE9
 
 | | High-pressure structure of pseudo-WT T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
2NTG
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R7 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(4-bromo-1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2OJL
 
 | | Crystal structure of Q7WAF1_BORPA from Bordetella parapertussis. Northeast Structural Genomics target BpR68. | | Descriptor: | Hypothetical protein | | Authors: | Benach, J, Neely, H, Seetharaman, J, Wang, H, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-12 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Q7WAF1_BORPA from Bordetella parapertussis
To be Published
|
|
2HFB
 
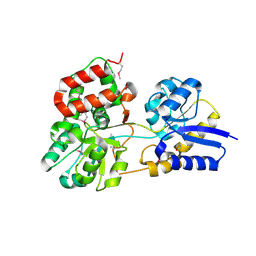 | |
4DEJ
 
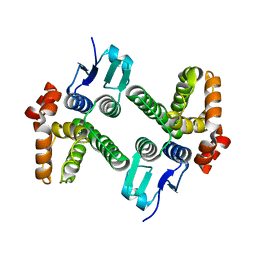 | | Crystal structure of glutathione transferase-like protein IL0419 (Target EFI-501089) from Idiomarina loihiensis L2TR | | Descriptor: | Glutathione S-transferase related protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-20 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase-Like Protein Il0419 from Idiomarina Loihiensis
To be Published
|
|
2OKW
 
 | |
4DGN
 
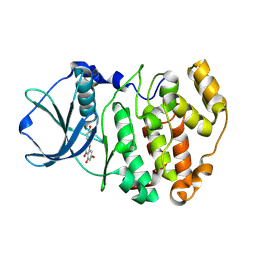 | | Crystal Structure of maize CK2 in complex with the inhibitor luteolin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Casein kinase II subunit alpha | | Authors: | Lolli, G, Mazzorana, M, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-08-01 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of protein kinase CK2 by flavonoids and tyrphostins. A structural insight.
Biochemistry, 51, 2012
|
|
2K5E
 
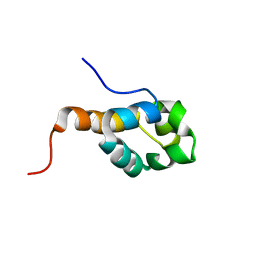 | | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195 | | Descriptor: | uncharacterized protein | | Authors: | Liu, G, Zhao, L, Ciccosanti, C, Jiang, M, Xiao, R, Swapna, G, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195
To be Published
|
|
4DHP
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
2OMV
 
 | | Crystal structure of InlA S192N Y369S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2OLZ
 
 | |
4DW3
 
 | | Crystal structure of the glycoprotein Erns from the pestivirus BVDV-1 in complex with 5'-CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Krey, T, Bontems, F, Vonrhein, C, Vaney, M.-C, Bricogne, G, Ruemenapf, T, Rey, F.A. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Pestivirus Envelope Glycoprotein E(rns) and Mechanistic Analysis of Its Ribonuclease Activity.
Structure, 20, 2012
|
|
4DWA
 
 | | Crystal structure of an active-site mutant of the glycoprotein Erns from the pestivirus BVDV-1 in complex with a CpUpC trinucleotide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E(rns) glycoprotein, ... | | Authors: | Krey, T, Bontems, F, Vonrhein, C, Vaney, M.-C, Bricogne, G, Ruemenapf, T, Rey, F.A. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of the Pestivirus Envelope Glycoprotein E(rns) and Mechanistic Analysis of Its Ribonuclease Activity.
Structure, 20, 2012
|
|
4DWF
 
 | |
2OMY
 
 | | Crystal structure of InlA S192N/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
4DYL
 
 | | F-BAR domain of human FES tyrosine kinase | | Descriptor: | Tyrosine-protein kinase Fes/Fps | | Authors: | Ugochukwu, E, Salah, E, Elkins, J, Barr, A, Krojer, T, Filippakopoulos, P, Weigelt, J, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | F-BAR domain of human FES tyrosine kinase
TO BE PUBLISHED
|
|
4D3G
 
 | | Structure of PstA | | Descriptor: | PSTA | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-10-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex Structure and Biochemical Characterization of the Staphylococcus Aureus Cyclic Di-AMP Binding Protein Psta, the Founding Member of a New Signal Transduction Protein Family
J.Biol.Chem., 290, 2015
|
|
2OMU
 
 | | Crystal structure of InlA G194S+S Y369S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OLV
 
 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L, Lim, D, Strynadka, N.C.J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
2OMW
 
 | | Crystal structure of InlA S192N Y369S/mEC1 complex | | Descriptor: | CHLORIDE ION, Epithelial-cadherin, Internalin-A | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2OQJ
 
 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
2K5I
 
 | | SOLUTION STRUCTURE OF IRON(II) TRANSPORT PROTEIN A FROM CLOSTRIDIUM THERMOCELLUM , NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET VR131 | | Descriptor: | IRON TRANSPORT PROTEIN | | Authors: | Liu, G, Wang, H, Foote, E.L, Jiang, M, Xiao, R, Swapna, G, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR chemical shift assignments of iron(ii) transport protein a from clostridium thermocellum , northeast structural genomics consortium (nesg) target vr131
To be Published
|
|
4CLV
 
 | | Crystal Structure of dodecylphosphocholine-solubilized NccX from Cupriavidus metallidurans 31A | | Descriptor: | NICKEL-COBALT-CADMIUM RESISTANCE PROTEIN NCCX, PHOSPHATE ION, PHOSPHOCHOLINE, ... | | Authors: | Legrand, P, Girard, E, Petit-Hartlein, I, Maillard, A.P, Coves, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The X-Ray Structure of Nccx from Cupriavidus Metallidurans 31A Illustrates Potential Dangers of Detergent Solubilization When Generating and Interpreting Crystal Structures of Membrane Proteins.
J.Biol.Chem., 289, 2014
|
|
4D02
 
 | | The crystallographic structure of Flavorubredoxin from Escherichia coli | | Descriptor: | ANAEROBIC NITRIC OXIDE REDUCTASE FLAVORUBREDOXIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Romao, C.V, Vicente, J.B, Bandeiras, T, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2014-04-23 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structure of Escherichia Coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
