5KY8
 
 | |
5L8N
 
 | | crystal structure of human FABP6 protein with fragment 1 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 5,6-dimethyl-1~{H}-benzimidazol-2-amine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
6PZS
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with JR005 | | Descriptor: | 4-[({[(1R,2R,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl]methyl}amino)methyl]-N-hydroxybenzamide, CHLORIDE ION, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-08-01 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Exploring Structural Determinants of Inhibitor Affinity and Selectivity in Complexes with Histone Deacetylase 6.
J.Med.Chem., 63, 2020
|
|
6D18
 
 | | Crystal structure of KPC-2 complexed with compound 6 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, [(5,7-dimethyl-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
6PYE
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NR160 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-[(1-benzyl-1H-tetrazol-5-yl)methyl]-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-2-(trifluoromethyl)benzamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-07-29 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.480003 Å) | | Cite: | Multicomponent Synthesis, Binding Mode, and Structure-Activity Relationship of Selective Histone Deacetylase 6 (HDAC6) Inhibitors with Bifurcated Capping Groups.
J.Med.Chem., 63, 2020
|
|
6Q74
 
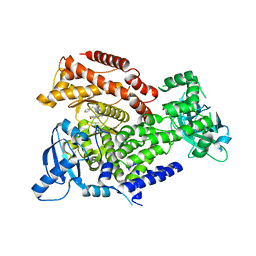 | | PI3K delta in complex with 1benzylN[5(3,6dihydro2Hpyran4yl)2methoxypyridin3yl]2methyl1Himidazole4sulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[5-(3,6-dihydro-2~{H}-pyran-4-yl)-2-methoxy-pyridin-3-yl]-2-methyl-1-(phenylmethyl)imidazole-4-sulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
7X8K
 
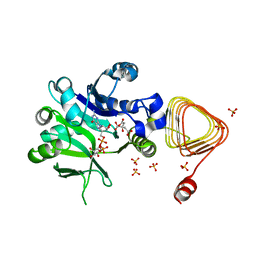 | | Arabidopsis GDP-D-mannose pyrophosphorylase (VTC1) structure (product-bound) | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, Mannose-1-phosphate guanylyltransferase 1, ... | | Authors: | Zhao, S, Zhang, C, Liu, L. | | Deposit date: | 2022-03-13 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana GDP-D-Mannose Pyrophosphorylase VITAMIN C DEFECTIVE 1.
Front Plant Sci, 13, 2022
|
|
4V3U
 
 | | Structure of human nNOS R354A G357D mutant heme domain in complex with N-2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)ethyl-3-(pyridin-3-yl) propan-1-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}-3-(pyridin-3-yl)propan-1-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
7WW7
 
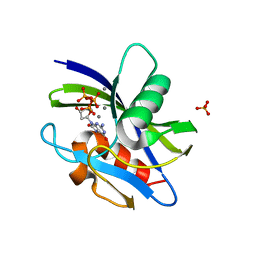 | | Crystal structure of MutT-8-oxo-dGTP complex: Reaction for 1 hr in 5 mM Mn2+ | | Descriptor: | 7,8-dihydro-8-oxoguanine-triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualization of mutagenic nucleotide processing by Escherichia coli MutT, a Nudix hydrolase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KO1
 
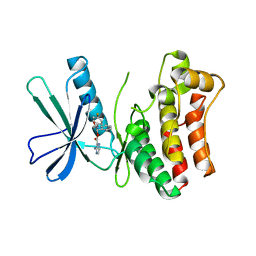 | | Pseudokinase Domain of MLKL bound to Compound 4. | | Descriptor: | Mixed lineage kinase domain-like protein, [(1~{R})-2-[(4-fluorophenyl)amino]-2-oxidanylidene-1-phenyl-ethyl] 3-azanylpyrazine-2-carboxylate | | Authors: | Marcotte, D.J. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | ATP-Competitive MLKL Binders Have No Functional Impact on Necroptosis.
Plos One, 11, 2016
|
|
6CBM
 
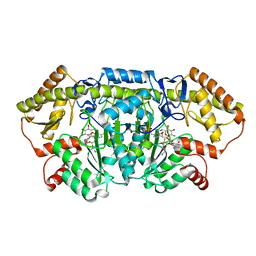 | | x-ray structure of NeoB from streptomyces fradiae in complex with PLP and neomycin (as the external aldimine) at pH 9 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-[(3-O-{2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranosyl}-beta-D-ribofuranosyl)oxy]-3-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Dow, G.T, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
3ZZF
 
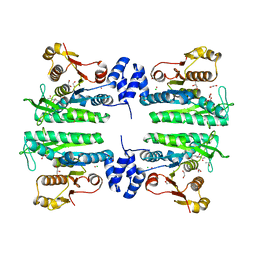 | | Crystal structure of the amino acid kinase domain from Saccharomyces cerevisiae acetylglutamate kinase complexed with its substrate N- acetylglutamate | | Descriptor: | 1,2-ETHANEDIOL, ACETYLGLUTAMATE KINASE, CHLORIDE ION, ... | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
2HR2
 
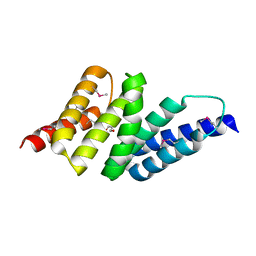 | |
1LA2
 
 | | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase | | Descriptor: | Myo-inositol-1-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kniewel, R, Buglino, J.A, Shen, V, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
2HP0
 
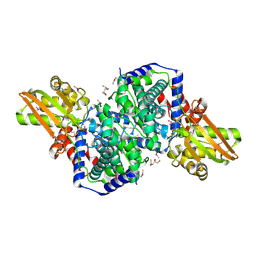 | | Crystal structure of iminodisuccinate epimerase | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, IDS-epimerase, ... | | Authors: | Lohkamp, B, Bauerle, B, Rieger, P.G, Schneider, G. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional Structure of Iminodisuccinate Epimerase Defines the Fold of the MmgE/PrpD Protein Family.
J.Mol.Biol., 362, 2006
|
|
2HTB
 
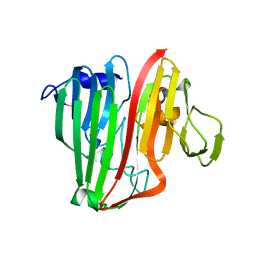 | | Crystal Structure of a putative mutarotase (YeaD) from Salmonella typhimurium in monoclinic form | | Descriptor: | Putative enzyme related to aldose 1-epimerase | | Authors: | Chittori, S, Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the putative mutarotase YeaD from Salmonella typhimurium: structural comparison with galactose mutarotases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5DR6
 
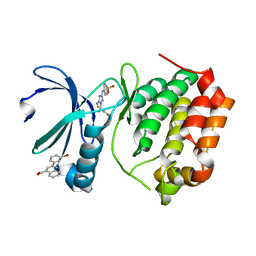 | | Aurora A Kinase in Complex with AA30 and JNJ-7706621 in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)quinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
5DRZ
 
 | | Crystal structure of anti-HIV-1 antibody F240 Fab in complex with gp41 peptide | | Descriptor: | Envelope glycoprotein gp160, HIV Antibody F240 Heavy Chain, HIV Antibody F240 Light Chain, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Sci Rep, 6, 2016
|
|
2HUM
 
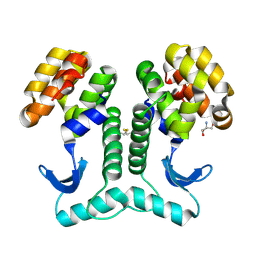 | |
5DT7
 
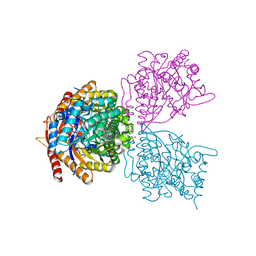 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group C2221 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
7WCL
 
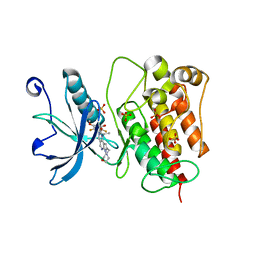 | | Crystal structure of FGFR1 kinase domain with Pemigatinib | | Descriptor: | 11-[2,6-bis(fluoranyl)-3,5-dimethoxy-phenyl]-13-ethyl-4-(morpholin-4-ylmethyl)-5,7,11,13-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),2(6),3,7-tetraen-12-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Lin, Q.M, Jiang, L.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Characterization of the cholangiocarcinoma drug pemigatinib against FGFR gatekeeper mutants.
Commun Chem, 5, 2022
|
|
6PDN
 
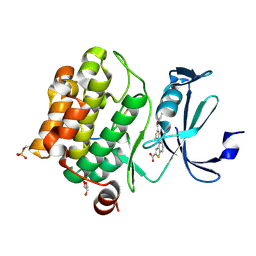 | | Human PIM1 bound to benzothiophene inhibitor 292 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxylic acid, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
5KKJ
 
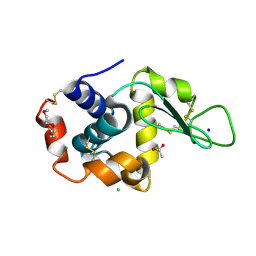 | | 2.0-Angstrom In situ Mylar structure of hen egg-white lysozyme (HEWL) at 293 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
5KKO
 
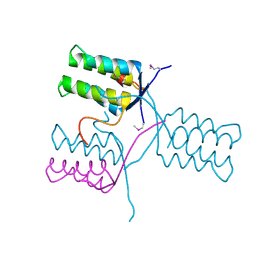 | | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein | | Descriptor: | Uncharacterised protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-22 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein
To Be Published
|
|
2I2F
 
 | | Crystal structure of LmNADK1 | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | Deposit date: | 2006-08-16 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
