7P93
 
 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus in complex with a sulfated ACKR1 N-terminal peptide | | Descriptor: | Atypical chemokine receptor 1, Leucotoxin LukEv | | Authors: | Lambey, P, Hoh, F, Peysson, F, Granier, S, Leyrat, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
6UI7
 
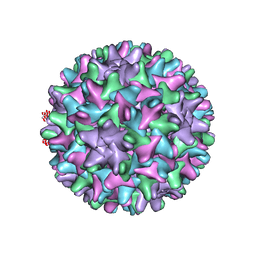 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
5GI4
 
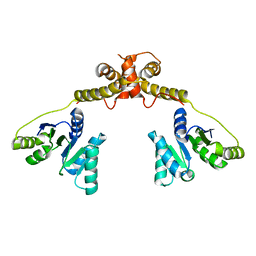 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
2J80
 
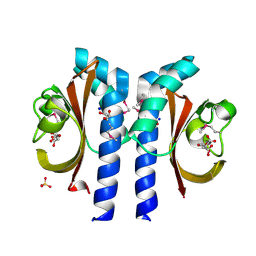 | | Structure of Citrate-bound Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | CITRATE ANION, GLYCEROL, SENSOR KINASE CITA, ... | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
7P8S
 
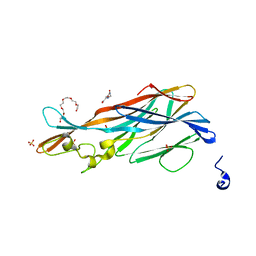 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus at 1.9 Angstrom resolution | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, HEXAETHYLENE GLYCOL, Leucotoxin LukEv, ... | | Authors: | Lambey, P, Hoh, F, Granier, S, Leyrat, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
9EAH
 
 | |
2D37
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
9EAJ
 
 | |
5C20
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound 2 | | Descriptor: | 2-methylpropyl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
2Y8K
 
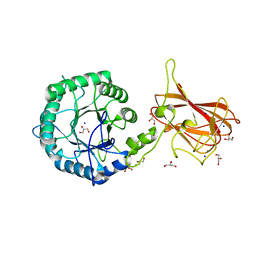 | | Structure of CtGH5-CBM6, an arabinoxylan-specific xylanase. | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Firbank, S.J, Correia, M.A, Mazumder, K, Bras, J.L, Zhu, Y, Lewis, R.J, York, W.S, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure and Function of an Arabinoxylan-Specific Xylanase.
J.Biol.Chem., 286, 2011
|
|
6LAB
 
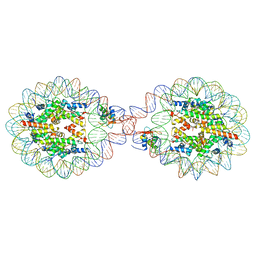 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
2IV7
 
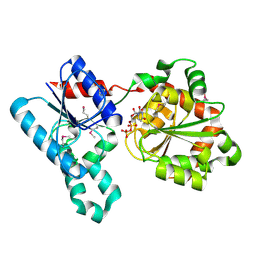 | | Crystal Structure of WaaG, a glycosyltransferase involved in lipopolysaccharide biosynthesis | | Descriptor: | LIPOPOLYSACCHARIDE CORE BIOSYNTHESIS PROTEIN RFAG, URIDINE-5'-DIPHOSPHATE | | Authors: | Martinez-Fleites, C, Proctor, M, Roberts, S, Bolam, D.N, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2006-06-08 | | Release date: | 2006-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights Into the Synthesis of Lipopolysaccharide and Antibiotics Through the Structures of Two Retaining Glycosyltransferases from Family Gt4
Chem.Biol., 13, 2006
|
|
5C2B
 
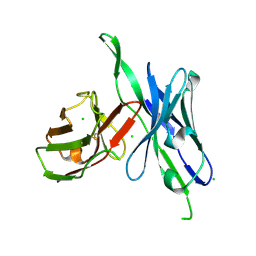 | | anti-CXCL13 parental scFv - 3B4 | | Descriptor: | CHLORIDE ION, scFv 3B4 | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-15 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4049 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
2IUY
 
 | | Crystal structure of AviGT4, a glycosyltransferase involved in Avilamycin A biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCOSYLTRANSFERASE, SULFATE ION | | Authors: | Martinez-Fleites, C, Proctor, M, Roberts, S, Bolam, D.N, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2006-06-08 | | Release date: | 2006-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Synthesis of Lipopolysaccharide and Antibiotics Through the Structures of Two Retaining Glycosyltransferases from Family Gt4
Chem.Biol., 13, 2006
|
|
6LER
 
 | | 169 bp nucleosome harboring non-identical cohesive DNA termini. | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Sharma, D, Adhireksan, Z, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-26 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
3GUY
 
 | | Crystal structure of a short-chain dehydrogenase/reductase from Vibrio parahaemolyticus | | Descriptor: | Short-chain dehydrogenase/reductase SDR | | Authors: | Patskovsky, Y, Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase from Vibrio parahaemolyticus
To be Published
|
|
8HHT
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Hit-1 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | | Authors: | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2022-11-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
1EXS
 
 | | STRUCTURE OF PORCINE BETA-LACTOGLOBULIN | | Descriptor: | BETA-LACTOGLOBULIN, GLYCEROL, SODIUM ION | | Authors: | Abrahams, J.P, Hoedemaeker, F.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A novel pH-dependent dimerization motif in beta-lactoglobulin from pig (Sus scrofa).
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8GY1
 
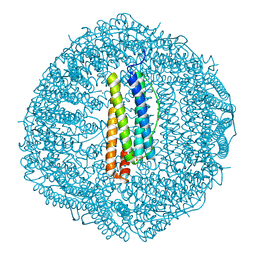 | |
8HCT
 
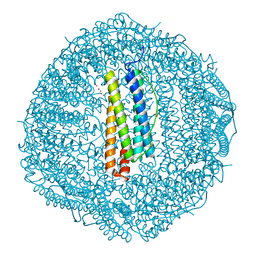 | | Crystal structure of Cu2+ binding to Dendrorhynchus zhejiangensis ferritin | | Descriptor: | COPPER (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Ming, T.H, Su, X.R, Huo, C.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Biochemical Characterization of Silver/Copper Binding by Dendrorhynchus zhejiangensis Ferritin.
Polymers (Basel), 15, 2023
|
|
9EAI
 
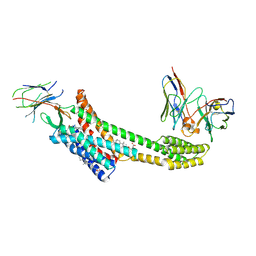 | | Structure of nanobody AT206 in complex with the losartan-bound angiotensin II type I receptor (AT1R) | | Descriptor: | BAG2 Anti-BRIL Fab Heavy Chain, BAG2 Anti-BRIL Fab Light Chain, CHOLESTEROL, ... | | Authors: | Skiba, M.A, Liu, J, Kruse, A.C. | | Deposit date: | 2024-11-11 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Epitope-directed selection of GPCR nanobody ligands with evolvable function.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
2YGJ
 
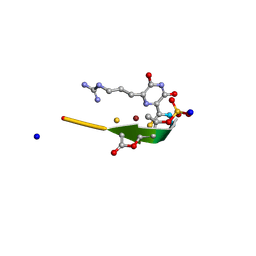 | | Methanobactin MB4 | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB4, SODIUM ION | | Authors: | Ghazouani, A, Basle, A, Firbank, S.J, Gray, J, Dennison, C. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Variations in Methanobactin Structure Influences Copper Utilization by Methane-Oxidizing Bacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2IUW
 
 | | Crystal structure of human ABH3 in complex with iron ion and 2- oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ALKYLATED REPAIR PROTEIN ALKB HOMOLOG 3, BETA-MERCAPTOETHANOL, ... | | Authors: | Sundheim, O, Vagbo, C.B, Bjoras, M, deSousa, M.M.L, Talstad, V, Aas, P.A, Drablos, F, Krokan, H.E, Tainer, J.A, Slupphaug, G. | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human Abh3 Structure and Key Residues for Oxidative Demethylation to Reverse DNA/RNA Damage.
Embo J., 25, 2006
|
|
3OY4
 
 | | Crystal Structure of HIV-1 L76V Protease in Complex with the Protease Inhibitor Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 PROTEASE, ... | | Authors: | Schiffer, C.A, Nalivaika, E.A, Bandaranayake, R.M. | | Deposit date: | 2010-09-22 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of HIV-1 L76V Protease in Complex with the Protease Inhibitor Darunavir.
To be Published
|
|
6U0I
 
 | |
