9F34
 
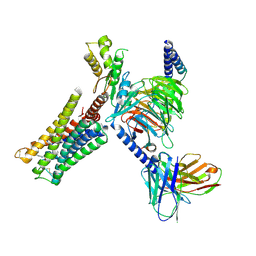 | |
9F33
 
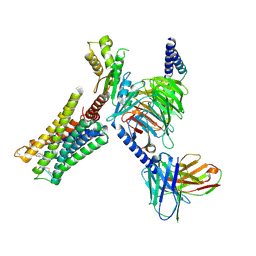 | |
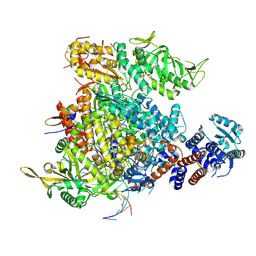
9F2R
 
 | |
9F2K
 
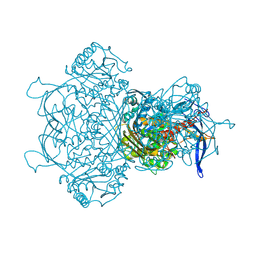 | | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, inositol-3-phosphate synthase | | Authors: | Traeger, T.K, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Disorder-to-order active site capping regulates the rate-limiting step of the inositol pathway.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9F1T
 
 | |
9F1S
 
 | |
9F1N
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46 | | Descriptor: | (2S,4R)-1-[(2S)-2-[9-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]nonanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-4H-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.71000016 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 46
to be published
|
|
9F1M
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-[7-[2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanoylamino]heptanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.90001261 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 45
to be published
|
|
9F1L
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 35 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[2-[2-[(3R)-2,6-bis(oxidanylidene)piperidin-3-yl]-1,3-bis(oxidanylidene)isoindol-4-yl]oxyethyl]-2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]ethanamide | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.30000627 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 35
to be published
|
|
9F1K
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]-N-(2-methoxyethyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30
to be published
|
|
9F1J
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[2-[2-[2-[2-[2-[2-[2,6-bis(oxidanylidene)piperidin-3-yl]-1,3-bis(oxidanylidene)isoindol-4-yl]oxyethanoylamino]ethoxy]ethoxy]ethoxy]ethyl]-4-[4-[2-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(2-morpholin-4-ylethyl)benzimidazol-2-yl]ethyl]phenoxy]butanamide | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.13003039 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based degrader 14
to be published
|
|
9F1I
 
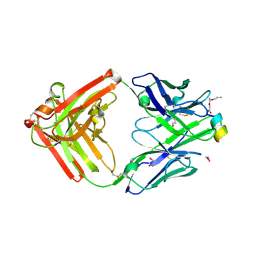 | | Crystal structure of a first-in-class antibody for alpha-1,6-fucosylated prostate-specific antigen, target bound | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-azanylethoxy)ethoxy]ethanoic acid, Heavy chain rabbit fab, ... | | Authors: | Halldorsson, S. | | Deposit date: | 2024-04-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Development of a first-in-class antibody and a specific assay for alpha-1,6-fucosylated prostate-specific antigen.
Sci Rep, 14, 2024
|
|
9F1G
 
 | |
9F1F
 
 | |
9F1E
 
 | |
9F1D
 
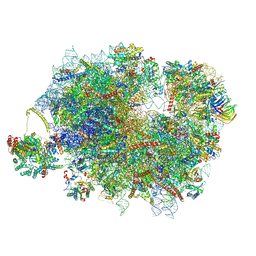 | | Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E-HYPK | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 2024
|
|
9F1C
 
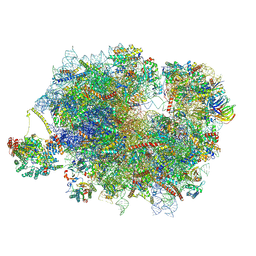 | | Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 2024
|
|
9F1B
 
 | | Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 2024
|
|
9F18
 
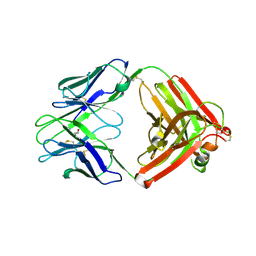 | |
9F17
 
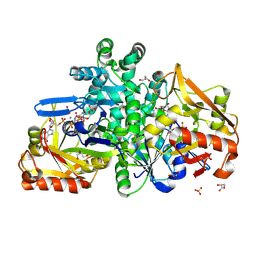 | |
9F14
 
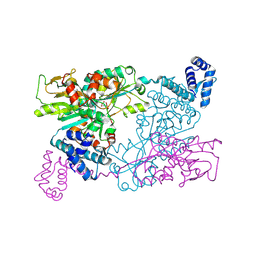 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
9F0G
 
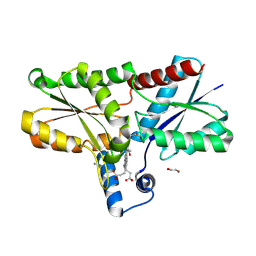 | | LmCpfC H182A variant in complex with iron coproporhyrin III | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, CALCIUM ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2024-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | LmCpfC H182A variant in complex with iron coproporhyrin III
To Be Published
|
|
9F0F
 
 | |
9F0E
 
 | |
9EZZ
 
 | | Bacterial histone protein HBb from Bdellovibrio bacteriovorus bound to DNA | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hu, Y, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2024-04-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacterial histone HBb from Bdellovibrio bacteriovorus compacts DNA by bending.
Nucleic Acids Res., 52, 2024
|
|
