6TCH
 
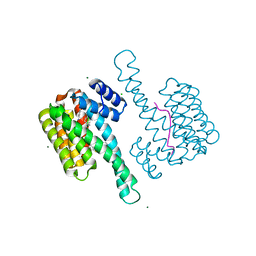 | |
5NCR
 
 | | OH1 from the Orf virus: a tyrosine phosphatase that displays distinct structural features and triple substrate specificity | | Descriptor: | PHOSPHATE ION, SULFATE ION, tyrosine phosphatase | | Authors: | Segovia, D, Haouz, A, Berois, M, Villarino, A, Andre-Leroux, G. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | OH1 from Orf Virus: A New Tyrosine Phosphatase that Displays Distinct Structural Features and Triple Substrate Specificity.
J. Mol. Biol., 429, 2017
|
|
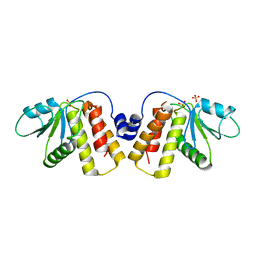
5H2P
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
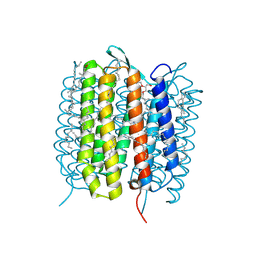
5H54
 
 | | Mdm12 from K. lactis 1-239 | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
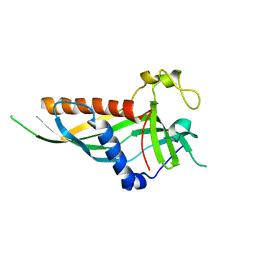
5N1C
 
 | |
5GVB
 
 | | SepB domain of human AND-1 | | Descriptor: | WD repeat and HMG-box DNA-binding protein 1 | | Authors: | Guan, C.C, Li, J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-04-19 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome.
J. Biol. Chem., 292, 2017
|
|
5N5W
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV3 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyaniline, CALCIUM ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
5GLW
 
 | | Tl-gal with LacNAc | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin | | Authors: | Jang, S.B, Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
6JLF
 
 | |
6JEZ
 
 | | Covalent labeling of rVDR-LBD by turn-on fluorescent probe mediated by conjugate addition and cyclization | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, 7-(diethylamino)chromen-2-one, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
5GMM
 
 | | Crystal structure of human Carbonic anhydrase I in complex with polmacoxib | | Descriptor: | 4-[3-(3-fluorophenyl)-5,5-dimethyl-4-oxidanylidene-furan-2-yl]benzenesulfonamide, Carbonic anhydrase 1, ZINC ION | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insight into the inhibition of carbonic anhydrase by the COX-2-selective inhibitor polmacoxib (CG100649).
Biochem. Biophys. Res. Commun., 478, 2016
|
|
6TOM
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3~{S},5~{R})-4,4-bis(fluoranyl)-3,5-dimethyl-piperidin-1-yl]-5-chloranyl-pyrimidin-4-yl]amino]-1-methyl-3-(3-methyl-3-oxidanyl-butyl)benzimidazol-2-one, B-cell lymphoma 6 protein | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
5H2H
 
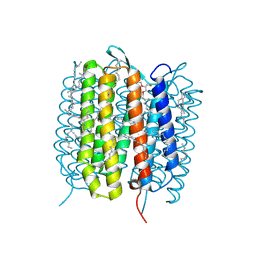 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 40 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5NCL
 
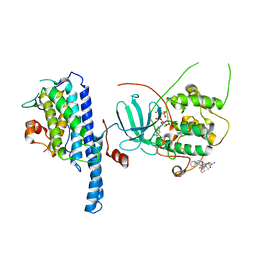 | | Crystal structure of the Cbk1-Mob2 kinase-coactivator complex with an SSD1 peptide | | Descriptor: | CBK1 kinase activator protein MOB2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein SSD1, ... | | Authors: | Gogl, G, Remenyi, A, Parker, B, Weiss, E. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Ndr/Lats Kinases Bind Specific Mob-Family Coactivators through a Conserved and Modular Interface.
Biochemistry, 2020
|
|
6JHB
 
 | |
5GKY
 
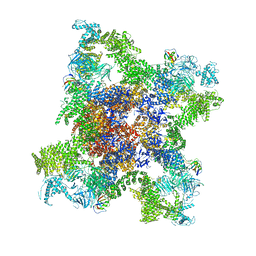 | | Structure of RyR1 in a closed state (C1 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
7BBD
 
 | | Crystal structure of monoubiquitinated TRIM21 RING (Ub-RING) In complex with ubiquitin charged Ube2N (Ube2N~Ub) and Ube2V2 | | Descriptor: | Polyubiquitin-B,E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Kiss, L, Neuhaus, D, James, L.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination.
Nat Commun, 12, 2021
|
|
6T38
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6T3Y
 
 | | Improved High Resolution Structure of MHC Class II complex | | Descriptor: | ACETATE ION, GLYCEROL, MHC class II alpha chain, ... | | Authors: | Halabi, S, Moncrieffe, M.C, Kaufman, J. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dominantly expressed class II molecule from a resistant MHC haplotype presents only a few Marek's disease virus peptides by using an unprecedented binding motif.
Plos Biol., 19, 2021
|
|
5GRH
 
 | |
6HFO
 
 | | Human Hsp90 co-chaperone TTC4 C-domain | | Descriptor: | PHOSPHATE ION, Tetratricopeptide repeat protein 4 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Co-chaperone Cns1 and the Recruiter Protein Hgh1 Link Hsp90 to Translation Elongation via Chaperoning Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
5H3T
 
 | | m7G cap bound to GEMIN5-WD | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, Gem-associated protein 5 | | Authors: | Bharath, S.R, Tang, X, Song, H. | | Deposit date: | 2016-10-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.571 Å) | | Cite: | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
7BVY
 
 | |
7BXZ
 
 | |
