3CL1
 
 | | M. loti cyclic-nucleotide binding domain, cyclic-GMP bound | | Descriptor: | CHLORIDE ION, CYCLIC GUANOSINE MONOPHOSPHATE, Mll3241 protein, ... | | Authors: | Clayton, G.M, Alteiri, S.L, Thomas, L.R, Morais-Cabral, J.H. | | Deposit date: | 2008-03-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Energetic Analysis of Activation by a Cyclic Nucleotide Binding Domain.
J.Mol.Biol., 381, 2008
|
|
6HQG
 
 | | Cytochrome P450-153 from Phenylobacterium zucineum | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
5DGX
 
 | | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
5DH3
 
 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
6U96
 
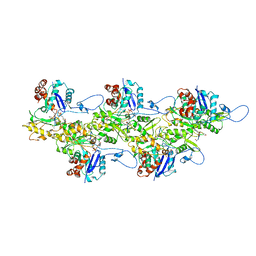 | | Actin phalloidin at BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Das, S, Ge, P, Durer, Z.A.O, Grintsevich, E.E, Zhou, Z.H, Reisler, E. | | Deposit date: | 2019-09-06 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | D-loop Dynamics and Near-Atomic-Resolution Cryo-EM Structure of Phalloidin-Bound F-Actin.
Structure, 28, 2020
|
|
8ALK
 
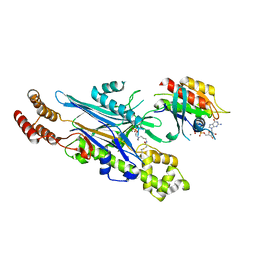 | | Structure of the Legionella phosphocholine hydrolase Lem3 in complex with its substrate Rab1 | | Descriptor: | 2-[[[5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxyethyl-[3-(2-chloranylethanoylamino)propyl]-dimethyl-azanium, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kaspers, M.S, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
5CPP
 
 | |
5CYX
 
 | |
3CYX
 
 | | Crystal structure of HIV-1 mutant I50V and inhibitor saquinavira | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
5D14
 
 | |
3CQP
 
 | | Human SOD1 G85R Variant, Structure I | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cao, X, Antonyuk, S, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
3CR6
 
 | |
6HWU
 
 | | Crystal structure of p38alpha in complex with a photoswitchable 2-Azothiazol-based Inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
5D1I
 
 | |
6HX4
 
 | | Fab fragment of a native monomer-selective antibody in complex with alpha-1-antitrypsin | | Descriptor: | Alpha-1-antitrypsin, Fab 1D9 heavy chain, Fab 1D9 light chain | | Authors: | Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Characterisation of a monoclonal antibody conformationally-selective for native alpha-1-antitrypsin
To Be Published
|
|
6HBJ
 
 | | Echovirus 18 empty particle | | Descriptor: | Viral protein 1, Viral protein 2, Viral protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
3CKW
 
 | | Crystal structure of sterile 20-like kinase 3 (MST3, STK24) | | Descriptor: | MERCURY (II) ION, Serine/threonine-protein kinase 24 | | Authors: | Antonysamy, S.S, Burley, S.K, Buchanan, S, Chau, F, Feil, I, Wu, L, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-17 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of sterile 20-like kinase 3 (MST3, STK24).
To be Published
|
|
8ATK
 
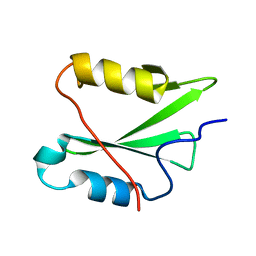 | | The SH2 domain of mouse SH2B1 | | Descriptor: | SH2B adapter protein 1 | | Authors: | Fowler, N.J, Williamson, M.P, Albalwi, M.F. | | Deposit date: | 2022-08-23 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Improved methodology for protein NMR structure calculation using hydrogen bond restraints and ANSURR validation: The SH2 domain of SH2B1.
Structure, 31, 2023
|
|
5DBQ
 
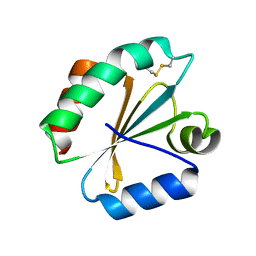 | | Crystal structure of insect thioredoxin at 1.95 Angstroms | | Descriptor: | Thioredoxin | | Authors: | Klinke, S, Tejedor, M.D, Cerutti, M.L, Giacometti, R, Otero, L.H, Goldbaum, F.A, Zavala, J.A, Wolosiuk, R.A, Pagano, E.A. | | Deposit date: | 2015-08-21 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of insect thioredoxin at 1.95 Angstroms
To Be Published
|
|
6UNI
 
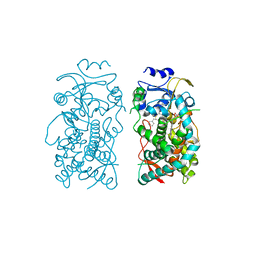 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
6HAB
 
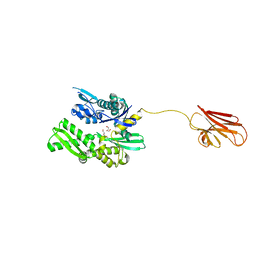 | | Crystal structure of BiP V461F (apo) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | MANF antagonizes nucleotide exchange by the endoplasmic reticulum chaperone BiP.
Nat Commun, 10, 2019
|
|
3CP5
 
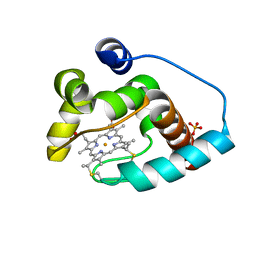 | | Cytochrome c from rhodothermus marinus | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Stelter, M, Melo, A, Saraiva, L, Teixeira, M, Archer, M. | | Deposit date: | 2008-03-31 | | Release date: | 2008-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A novel type of monoheme cytochrome c: biochemical and structural characterization at 1.23 A resolution of rhodothermus marinus cytochrome c
Biochemistry, 47, 2008
|
|
6HGF
 
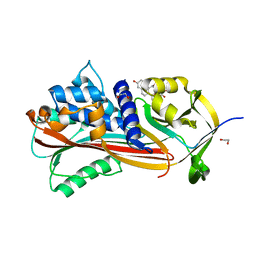 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-II: a new binding globulin in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
6ULB
 
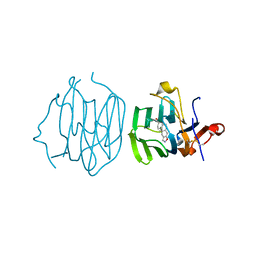 | |
