2GOV
 
 | | Solution structure of Murine p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Volkman, B.F, Dias, J.S, Goodfellow, B.J, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The First Structure from the SOUL/HBP Family of Heme-binding Proteins, Murine P22HBP.
J.Biol.Chem., 281, 2006
|
|
2GOW
 
 | | Solution structure of BC059385 from Homo sapiens | | Descriptor: | Ubiquitin-like protein 3 | | Authors: | Volkman, B.F, de la Cruz, N.B, Lytle, B.L, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-14 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a membrane-anchored ubiquitin-fold (MUB) protein from Homo sapiens.
Protein Sci., 16, 2007
|
|
2GOX
 
 | | Crystal structure of Efb-C / C3d Complex | | Descriptor: | Complement C3, Fibrinogen-binding protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-04-14 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural basis for complement inhibition by Staphylococcus aureus.
Nat.Immunol., 8, 2007
|
|
2GOY
 
 | | Crystal structure of assimilatory adenosine 5'-phosphosulfate reductase with bound APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, IRON/SULFUR CLUSTER, adenosine phosphosulfate reductase | | Authors: | Chartron, J, Carroll, K.S, Shiau, C, Gao, H, Leary, J.A, Bertozzi, C.R, Stout, C.D. | | Deposit date: | 2006-04-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate Recognition, Protein Dynamics, and Iron-Sulfur Cluster in Pseudomonas aeruginosa Adenosine 5'-Phosphosulfate Reductase.
J.Mol.Biol., 364, 2006
|
|
2GP0
 
 | | HePTP Catalytic Domain (residues 44-339), S225D mutant | | Descriptor: | GLYCEROL, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 7 | | Authors: | Page, R. | | Deposit date: | 2006-04-15 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | HePTP Catalytic Domain (residues 44-339), S225D mutant
To be Published
|
|
2GP1
 
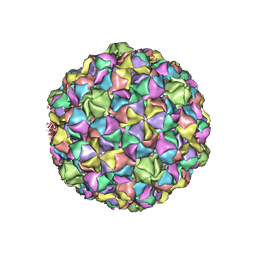 | |
2GP3
 
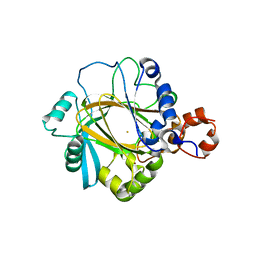 | | Crystal structure of the catalytic core domain of jmjd2a | | Descriptor: | FE (II) ION, Jumonji domain-containing protein 2A, ZINC ION | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
2GP4
 
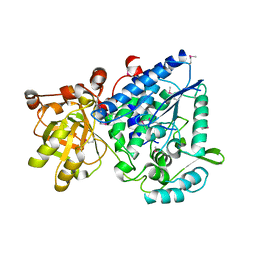 | |
2GP5
 
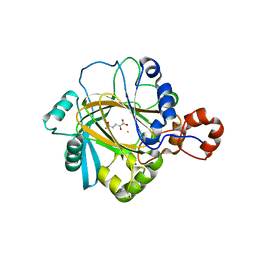 | | Crystal structure of catalytic core domain of jmjd2A complexed with alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Jumonji domain-containing protein 2A, ... | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
2GP6
 
 | |
2GP7
 
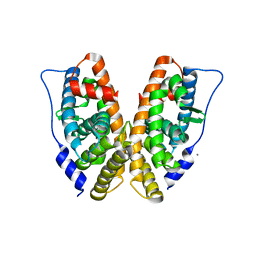 | | Estrogen Related Receptor-gamma ligand binding domain | | Descriptor: | CALCIUM ION, Estrogen-related receptor gamma | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-17 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
2GP8
 
 | | NMR SOLUTION STRUCTURE OF THE COAT PROTEIN-BINDING DOMAIN OF BACTERIOPHAGE P22 SCAFFOLDING PROTEIN | | Descriptor: | PROTEIN (SCAFFOLDING PROTEIN) | | Authors: | Sun, Y, Parker, M.H, Weigele, P, Casjens, S, Prevelige Jr, P.E, Krishna, N.R. | | Deposit date: | 1999-05-11 | | Release date: | 1999-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus.
J.Mol.Biol., 297, 2000
|
|
2GP9
 
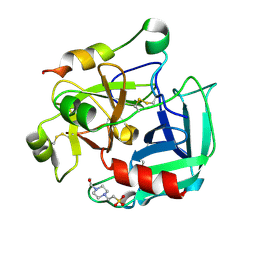 | | Crystal structure of the slow form of thrombin in a self-inhibited conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Prothrombin | | Authors: | Pineda, A, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-04-17 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of thrombin in a self-inhibited conformation.
J.Biol.Chem., 281, 2006
|
|
2GPA
 
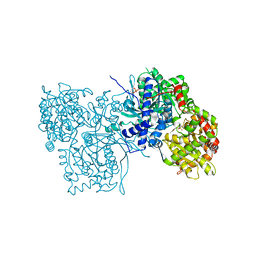 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
2GPB
 
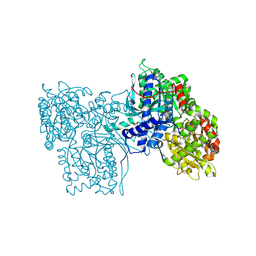 | |
2GPC
 
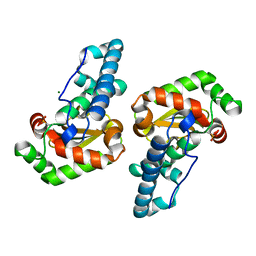 | |
2GPE
 
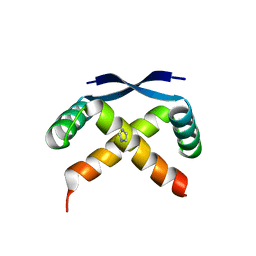 | |
2GPF
 
 | |
2GPH
 
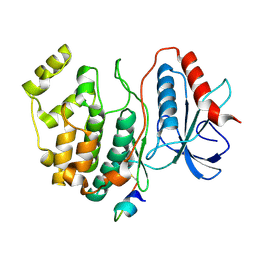 | | Docking motif interactions in the MAP kinase ERK2 | | Descriptor: | Mitogen-activated protein kinase 1, Tyrosine-protein phosphatase non-receptor type 7 | | Authors: | Zhou, T, Sun, L, Humphreys, J, Goldsmith, E.J. | | Deposit date: | 2006-04-17 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Docking Interactions Induce Exposure of Activation Loop in the MAP Kinase ERK2.
Structure, 14, 2006
|
|
2GPI
 
 | |
2GPJ
 
 | |
2GPL
 
 | | TMC-95 based biphenyl-ether macrocycles: specific proteasome inhibitors | | Descriptor: | BENZYL [12-(2-AMINO-2-OXOETHYL)-4-NITRO-10,13-DIOXO-15-[(PROPYLAMINO)CARBONYL]-2-OXA-11,14-DIAZATRICYCLO[15 .2.2.1~3,7~]DOCOSA-1(19),3(22),4,6,17,20-HEXAEN-9-YL]CARBAMATE, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Goetz, M, Kaiser, M, Weyher, E, Moroder, M. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | TMC-95-Based Inhibitor Design Provides Evidence for the Catalytic Versatility of the Proteasome.
Chem.Biol., 13, 2006
|
|
2GPM
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3') | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GPN
 
 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
2GPO
 
 | | Estrogen Related Receptor-gamma ligand binding domain complexed with a synthetic peptide from RIP140 | | Descriptor: | Estrogen-related receptor gamma, Nuclear receptor-interacting protein 1 | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-18 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
