8K5T
 
 | |
7DYS
 
 | |
8K8M
 
 | |
6V7T
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase in complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6VGG
 
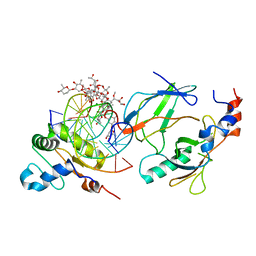 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), and mithramycin, in complex with 16mer DNA CAGAGGATGTGGCTTC | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2020-01-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
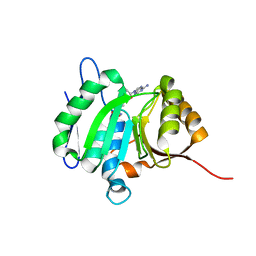
8HL6
 
 | |
6VBI
 
 | | crystal structure of PDE5 in complex with a non-competitive inhibitor | | Descriptor: | (13bS)-4,9-dimethoxy-14-methyl-8,13,13b,14-tetrahydroindolo[2',3':3,4]pyrido[2,1-b]quinazolin-5(7H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Ke, H, Luo, H.B. | | Deposit date: | 2019-12-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.30000758 Å) | | Cite: | Identification of a novel allosteric pocket and its regulation mechanism
To Be Published
|
|
8HL7
 
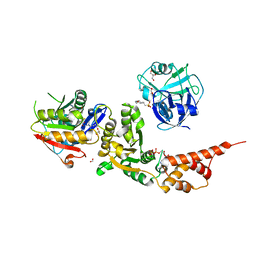 | |
6VG8
 
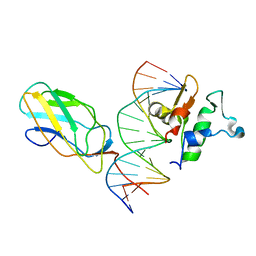 | |
8P5T
 
 | | Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
6VGE
 
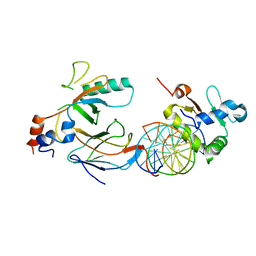 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), in complex with 16mer DNA CAGAGGATGTGGCTTC | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | Authors: | Hou, C, Tsodikov, O.V. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
8P5V
 
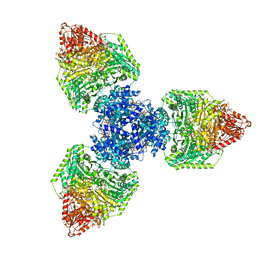 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5U
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum with Coenzyme A bound to the E2o domain | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5X
 
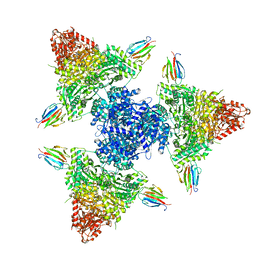 | | Single particle cryo-EM structure of the complex between Corynebacterium glutamicum homohexameric 2-oxoglutarate dehydrogenase OdhA and the FHA-protein inhibitor OdhI | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, MAGNESIUM ION, Oxoglutarate dehydrogenase inhibitor, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
6V4V
 
 | |
7E42
 
 | |
6URS
 
 | | Sleeping Beauty transposase PAI subdomain mutant - H19Y | | Descriptor: | Sleeping Beauty transposase PAI subdomain | | Authors: | Nesmelova, I.V, Leighton, G.O, Yan, C, Lustig, J, Corona, R.I, Guo, J.T, Ivics, Z. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | H19Y mutation in the primary DNA-recognition subdomain of the Sleeping Beauty transposase improves structural stability, transposon DNA-binding and transposition
To Be Published
|
|
6VIZ
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VKE
 
 | | Crystal Structure of Inhibitor JNJ-40012665 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(5-chloro-2-{[1-(3,4-dimethoxyphenyl)-2-oxo-1,2-dihydro-3H-imidazo[4,5-c]pyridin-3-yl]methyl}-1H-indol-1-yl)butanenitrile, CHLORIDE ION, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
8P5W
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum following reaction with the 2-oxoglutarate analogue succinyl phosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
6VOQ
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae | | Descriptor: | Aldolase, CHLORIDE ION, ZINC ION | | Authors: | Stogios, P.J, Evdokimova, E, McChesney, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae
To Be Published
|
|
6VSX
 
 | |
8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
6VPR
 
 | |
6VPX
 
 | |
