7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFC
 
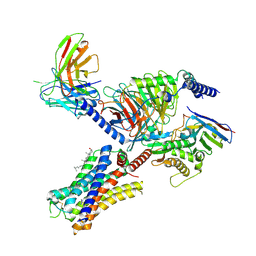 | | Cryo-EM structure of the histamine-bound histamine H4 receptor and Gq complex | | Descriptor: | CHOLESTEROL, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
7YFD
 
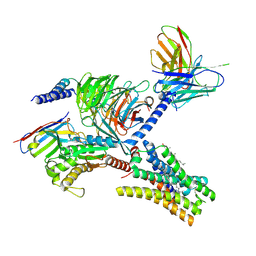 | | Cryo-EM structure of the imetit-bound histamine H4 receptor and Gq complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, CHOLESTEROL, Engineered G-alpha-q, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
7YF1
 
 | |
7YF6
 
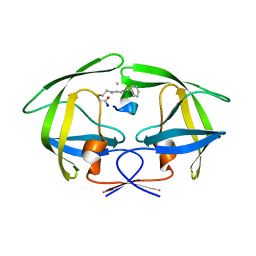 | | Crystal structure of HIV-1 protease in complex with macrocyclic peptide | | Descriptor: | MAGNESIUM ION, Macrocyclic Peptide, Protease | | Authors: | Kusumoto, Y, Sato, S, Yamada, T, Kozono, I, Nakata, Z, Asada, N, Mitsuki, S, Wakasa-Morimoto, C, Tohru, M, Watanabe, A, Hayashi, K, Mikamiyama, H. | | Deposit date: | 2022-07-07 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Highly Potent and Oral Macrocyclic Peptides as a HIV-1 Protease Inhibitor: mRNA Display-Derived Hit-to-Lead Optimization.
Acs Med.Chem.Lett., 13, 2022
|
|
8DKU
 
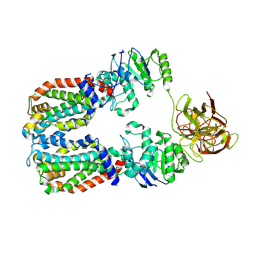 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, Transport permease protein | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
8DL0
 
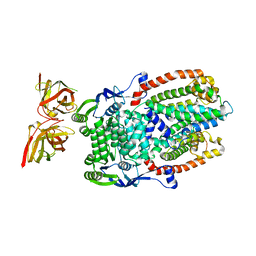 | |
8DL4
 
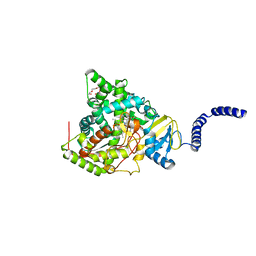 | | S. CEREVISIAE CYP51 COMPLEXED WITH Courmarin-containing INHIBITOR | | Descriptor: | 7-(diethylamino)-N-[(2S)-2-(2,4-difluorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]-2-oxo-2H-1-benzopyran-3-carboxamide, Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, ... | | Authors: | Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | S. CEREVISIAE CYP51 COMPLEXED WITH Courmarin-containing INHIBITOR
To Be Published
|
|
8ACT
 
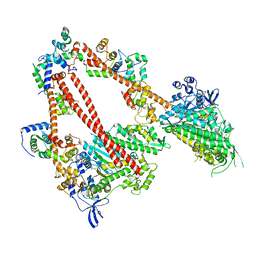 | | structure of the human beta-cardiac myosin folded-back off state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light chain 3, ... | | Authors: | Grinzato, A, Kandiah, E, Robert-Paganin, J, Auguin, D, Kikuti, C, Nandwani, N, Moussaoui, D, Pathak, D, Ruppel, K.M, Spudich, J.A, Houdusse, A. | | Deposit date: | 2022-07-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the folded-back state of human beta-cardiac myosin.
Nat Commun, 14, 2023
|
|
7YEY
 
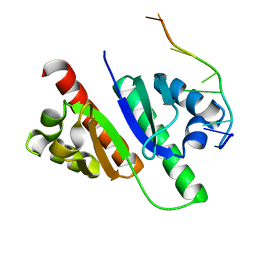 | | Structure of MmIGF2BP3-KH12 in complex with 8-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(*CP*AP*CP*GP*GP*CP*AP*C)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of MmIGF2BP3-KH12 in complex with 8-mer RNA
To Be Published
|
|
7YEX
 
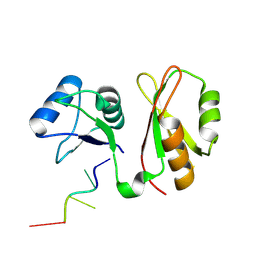 | | Structure of MmIGF2BP3 in complex with 8-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(P*CP*AP*CP*AP*CP*AP*CP*A)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of MmIGF2BP3 in complex with 8-mer RNA
To Be Published
|
|
7YEW
 
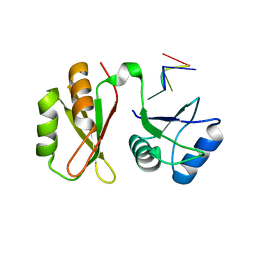 | | Structure of MmIGF2BP3 in complex with 7-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(P*CP*AP*CP*AP*CP*AP*C)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of MmIGF2BP3 in complex with 7-mer RNA
To Be Published
|
|
8AC5
 
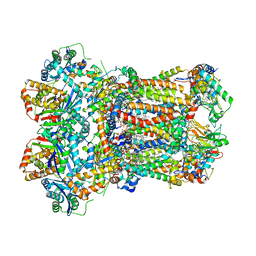 | | Complex III2 from Yarrowia lipolytica, with decylubiquinol, oxidised, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AC4
 
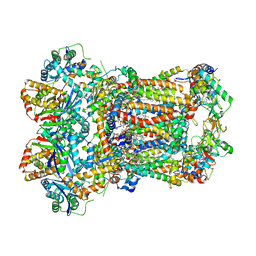 | | Complex III2 from Yarrowia lipolytica, apo, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AC3
 
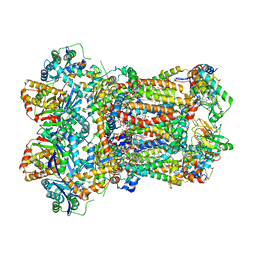 | | Complex III2 from Yarrowia lipolytica, apo, int-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ACM
 
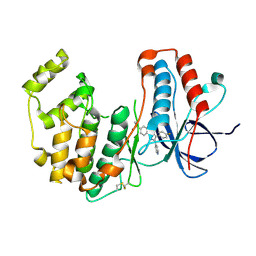 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACO
 
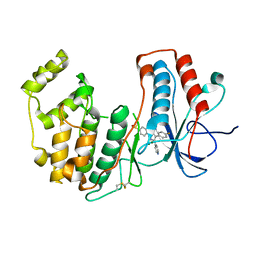 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACF
 
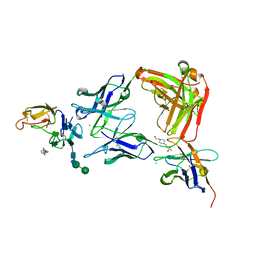 | |
8ACI
 
 | |
8AB3
 
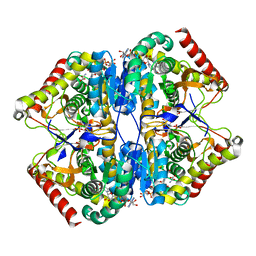 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8AB2
 
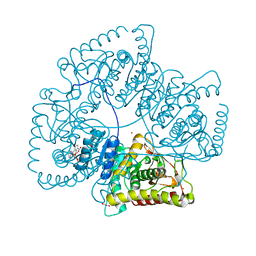 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8ABI
 
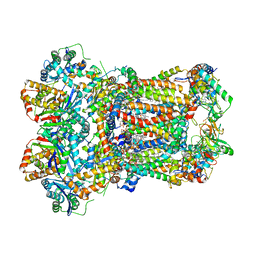 | | Complex III2 from Yarrowia lipolytica,antimycin A bound, int-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AB7
 
 | | Complex III2 from Yarrowia lipolytica, atovaquone and antimycin A bound | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2-[trans-4-(4-chlorophenyl)cyclohexyl]-3-hydroxynaphthalene-1,4-dione, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AB9
 
 | | Complex III2 from Yarrowia lipolytica, ascorbate-reduced, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ABB
 
 | | Complex III2 from Yarrowia lipolytica, ascorbate-reduced, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
