7UKK
 
 | |
7UJU
 
 | | Room-temperature X-ray structure of monomeric SARS-CoV-2 main protease catalytic domain (MPro1-196) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-03-31 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Autoprocessing and oxyanion loop reorganization upon GC373 and nirmatrelvir binding of monomeric SARS-CoV-2 main protease catalytic domain.
Commun Biol, 5, 2022
|
|
6ZAU
 
 | | Damage-free nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
5N2E
 
 | | Structure of the E9 DNA polymerase from vaccinia virus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tarbouriech, N, Burmeister, W.P, Iseni, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The vaccinia virus DNA polymerase structure provides insights into the mode of processivity factor binding.
Nat Commun, 8, 2017
|
|
8INJ
 
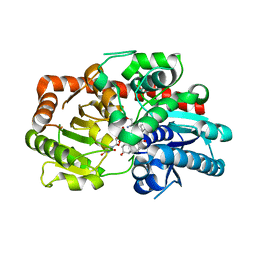 | | Crystal structure of UGT74AN3-UDP-DIG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | Authors: | Feng, L, Wei, H. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of UGT74AN3-UDP-DIG
To Be Published
|
|
8SAG
 
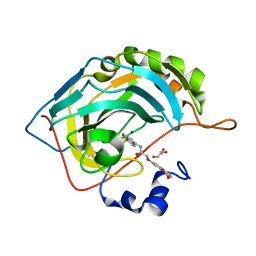 | |
6H7X
 
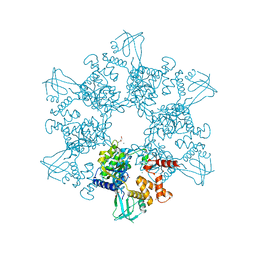 | | First X-ray structure of full-length human RuvB-Like 2. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Silva, S, Brito, J.A, Matias, P, Bandeiras, T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | X-ray structure of full-length human RuvB-Like 2 - mechanistic insights into coupling between ATP binding and mechanical action.
Sci Rep, 8, 2018
|
|
5MZY
 
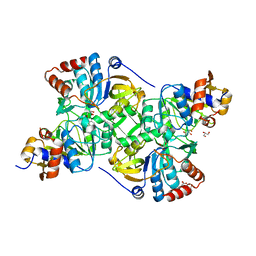 | | Crystal structure of the decarboxylase AibA/AibB in complex with a possible transition state analog | | Descriptor: | (1~{R},2~{S})-2-methylcyclohexane-1-carboxylic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6GN1
 
 | | Crystal Structure of Glycogen synthase kinase-3 beta (GSK3B) in Complex with PIK-75 | | Descriptor: | CHLORIDE ION, Glycogen synthase kinase-3 beta, ~{N}-[(~{E})-(6-bromanylimidazo[1,2-a]pyridin-3-yl)methylideneamino]-~{N},2-dimethyl-5-nitro-benzenesulfonamide | | Authors: | Tesch, R, Becker, C, Mueller, M.P, Sant'Anna, C.M.R, Fraga, C.A.M, Rauh, D. | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Unusual Intramolecular Halogen Bond Guides Conformational Selection.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
9BBB
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, cobicistat | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of CYP3A4 with the inhibitor cobicistat: Structural and mechanistic insights and comparison with ritonavir.
Arch.Biochem.Biophys., 758, 2024
|
|
5NFZ
 
 | | TUBULIN-MTC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-(2,3,4-trimethoxyphenyl)cyclohepta-2,4,6-trien-1-one, CALCIUM ION, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
8P9X
 
 | | Vitamin D receptor complex with Xe4Me agonist ligand | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-1,7~{a}-dimethyl-1-(5-methyl-5-oxidanyl-hexa-1,3-diynyl)-2,3,3~{a},5,6,7-hexahydroinden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A vitamin D-based strategy overcomes chemoresistance in prostate cancer.
Br.J.Pharmacol., 181, 2024
|
|
6RY1
 
 | |
8OVC
 
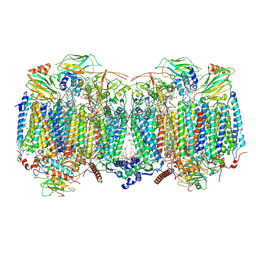 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
8X73
 
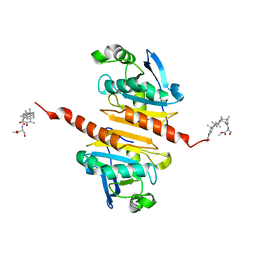 | | Crystal structure of Peroxiredoxin I in complex with compound 19-069 | | Descriptor: | Peroxiredoxin-1, methyl (2~{S})-2-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]-3-oxidanyl-propanoate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2023-11-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8X71
 
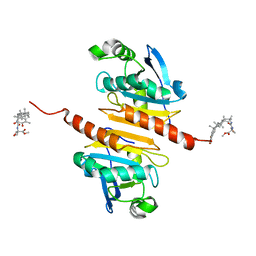 | | Crystal structure of Peroxiredoxin I in complex with compound 19-064 | | Descriptor: | Peroxiredoxin-1, methyl 3-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]oxetane-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2023-11-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8HYC
 
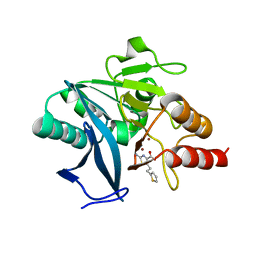 | |
6RZI
 
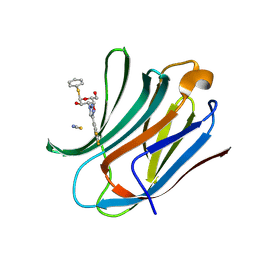 | | Galectin-3C in complex with trifluoroaryltriazole monothiogalactoside derivative:1(Hydrogen) | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-phenylsulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Verteramo, M.L, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Interplay of halogen bonding and solvation in protein-ligand binding.
Iscience, 27, 2024
|
|
5YHA
 
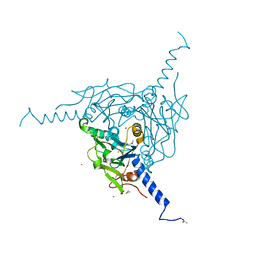 | | Crystal structure of Pd(allyl)/Wild Type Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PALLADIUM ION, ... | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
7UM8
 
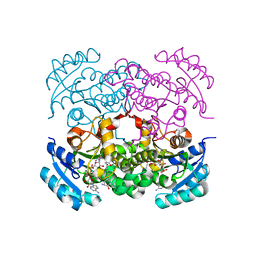 | | Crystal structure of E. Coli FabI in complex with NAD and (R,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide | | Descriptor: | (2E)-3-[(7R)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hajian, B. | | Deposit date: | 2022-04-06 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
5KJ0
 
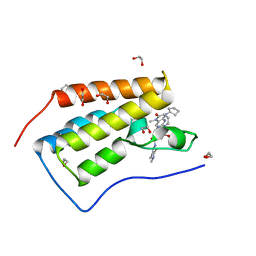 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH DB-1-264-2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(7~{R})-8-cyclopentyl-7-ethyl-5-methyl-6-oxidanylidene-7~{H}-pteridin-2-yl]-methyl-amino]-3-methoxy-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W, Schonbrunn, E. | | Deposit date: | 2016-06-17 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assessment of Bromodomain Target Engagement by a Series of BI2536 Analogues with Miniaturized BET-BRET.
ChemMedChem, 11, 2016
|
|
8ECG
 
 | | Complex of HMG1 with pitavastatin | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1, GLYCEROL, Pitavastatin | | Authors: | Haywood, J, Bond, C.S. | | Deposit date: | 2022-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.
Nat Commun, 13, 2022
|
|
5KRK
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-((5-bromo-2,3-dihydro-1H-inden-1-ylidene)methylene)diphenol | | Descriptor: | 4-[(5-bromanyl-2,3-dihydroinden-1-ylidene)-(4-hydroxyphenyl)methyl]phenol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
8S2M
 
 | |
5NK1
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1k | | Descriptor: | 2-[[3-[(4-azanyl-6-methyl-1,3,5-triazin-2-yl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
