8TXY
 
 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5M67
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenine and 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, ACETATE ION, ADENINE, ... | | Authors: | Manszewski, T, Jaskolski, M. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
5A46
 
 | | FGFR1 in complex with dovitinib | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
4FVL
 
 | | Human collagenase 3 (MMP-13) full form with peptides from pro-domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase 3, ... | | Authors: | Stura, E.A, Vera, L, Visse, R, Nagase, H, Dive, V. | | Deposit date: | 2012-06-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Crystal structure of full-length human collagenase 3 (MMP-13) with peptides in the active site defines exosites in the catalytic domain.
Faseb J., 27, 2013
|
|
3UFV
 
 | | Structure of rat nitric oxide synthase heme domain in complex with 4-methyl-6-(((3R,4R)-4-((5-(4-methylpyridin-2-yl)pentyl)oxy)pyrrolidin-3-yl)methyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-{[(3R,4R)-4-{[5-(4-methylpyridin-2-yl)pentyl]oxy}pyrrolidin-3-yl]methyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2011-11-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Selective monocationic inhibitors of neuronal nitric oxide synthase. Binding mode insights from molecular dynamics simulations.
J.Am.Chem.Soc., 134, 2012
|
|
5M7Z
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2) | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 4E, GLYCEROL, ... | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2)
To Be Published
|
|
3UFQ
 
 | | Structure of rat nitric oxide synthase heme domain in complex with 6-(((3S,4S)-4-(((E)-5-(3-fluorophenyl)pent-4-en-1-yl)oxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-{[(4E)-5-(3-fluorophenyl)pent-4-en-1-yl]oxy}pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2011-11-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.058 Å) | | Cite: | Selective monocationic inhibitors of neuronal nitric oxide synthase. Binding mode insights from molecular dynamics simulations.
J.Am.Chem.Soc., 134, 2012
|
|
4PZ3
 
 | |
2ZWF
 
 | |
8TGP
 
 | |
3UGP
 
 | |
5M5S
 
 | |
3UGU
 
 | |
1O1G
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle.
J.Struct.Biol., 138, 2002
|
|
8TPE
 
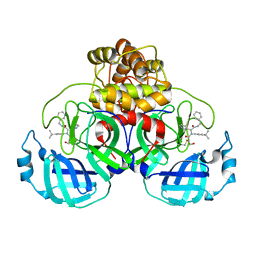 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPG
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
4G44
 
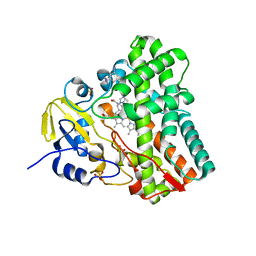 | | Structure of P450 CYP121 in complex with lead compound MB286, 3-((1H-1,2,4-triazol-1-yl)methyl)aniline | | Descriptor: | 3-(1H-1,2,4-triazol-1-ylmethyl)aniline, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5MC5
 
 | | Crystal Structure of delGlu452 mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, HYDROXIDE ION, ... | | Authors: | Wilk, P, Mueller, U, Dobbek, H, Weiss, M.S. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
5MCP
 
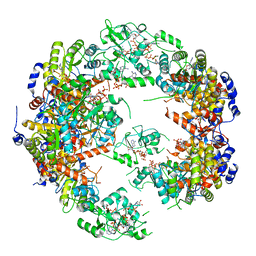 | | Structure of IMP dehydrogenase from Ashbya gossypii bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Winter, G, Fernandez-Justel, D, de Pereda, J.M, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep, 7, 2017
|
|
8TPF
 
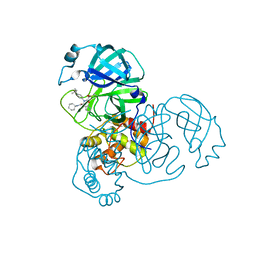 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
5M6B
 
 | |
5M75
 
 | |
8TPB
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-2-chloroacetamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
5M80
 
 | | Translation initiation factor 4E in complex with (RP)-iPr-m7GppSpG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Translation initiation factor 4E in complex with (RP)-iPr-m7GppSpG mRNA 5' cap analog
To Be Published
|
|
8TPH
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
