4I6B
 
 | |
4I6F
 
 | |
7WFF
 
 | | Subcomplexes B,M and L in the Cylic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFE
 
 | | Right PSI in the cyclic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFG
 
 | | Subcomplexes A and E in NDH complex from Arabidopsis | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase subunit H, chloroplastic, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WG5
 
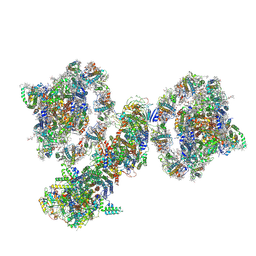 | | Cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFD
 
 | | Left PSI in the cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pan, X, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7X9Q
 
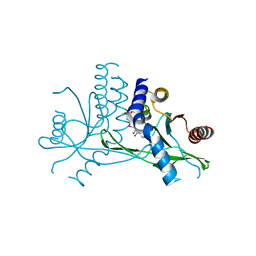 | | Crystal structure of human STING complexed with compound BSP16 | | Descriptor: | (2R)-4-(5,6-dimethoxy-1-benzoselenophen-2-yl)-2-ethyl-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Selenium-Containing STING Agonists as Orally Available Antitumor Agents.
J.Med.Chem., 65, 2022
|
|
7X9P
 
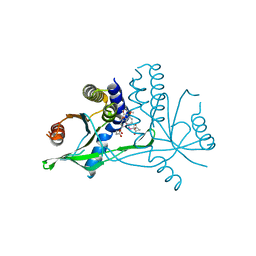 | | Crystal structure of human STING complexed with compound BSP17 | | Descriptor: | 4-[6-methoxy-5-[3-[[6-methoxy-2-(4-oxidanyl-4-oxidanylidene-butanoyl)-1-benzoselenophen-5-yl]oxy]propoxy]-1-benzoselenophen-2-yl]-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Selenium-containing STING agonists as orally available anti tumor agents
To be published
|
|
7DDX
 
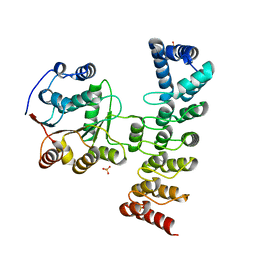 | | Crystal structure of KANK1 S1179F mutant in complex wtih eIF4A1 | | Descriptor: | Eukaryotic initiation factor 4A-I, GLYCEROL, KN motif and ankyrin repeat domains 1, ... | | Authors: | Pan, W, Xu, Y, Wei, Z. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nephrotic-syndrome-associated mutation of KANK2 induces pathologic binding competition with physiological interactor KIF21A.
J.Biol.Chem., 297, 2021
|
|
7KFV
 
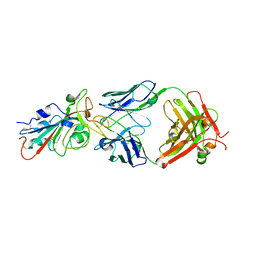 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B12 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody C1A-B12 Fab, Spike glycoprotein, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFY
 
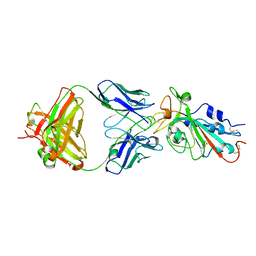 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-F10 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-F10 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFW
 
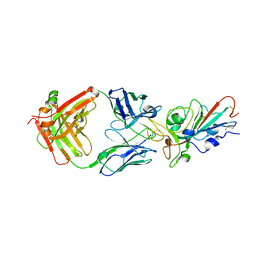 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B3 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of antibody C1A-B3 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFX
 
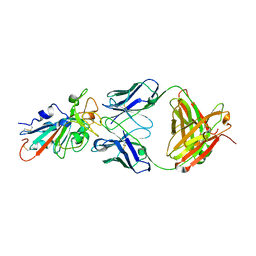 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-C2 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-C2 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
1EV0
 
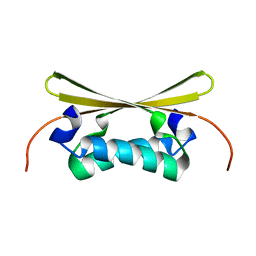 | |
1DJM
 
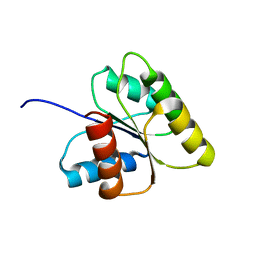 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
6OLD
 
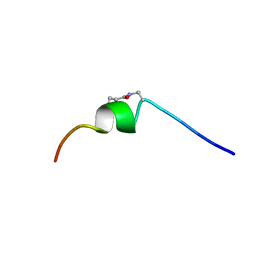 | | CSP1-cyc(Dap6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-04-16 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1CR9
 
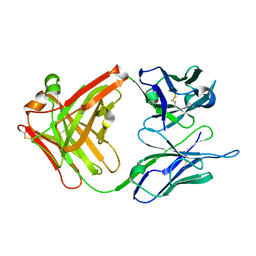 | | CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Kanyo, Z.F, Pan, K.M, Williamson, R.A, Burton, D.R, Prusiner, S.B, Fletterick, R.J, Cohen, F.E. | | Deposit date: | 1999-08-14 | | Release date: | 2000-04-17 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change.
J.Mol.Biol., 293, 1999
|
|
1CU4
 
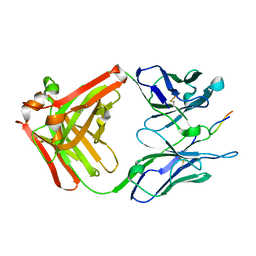 | | CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 IN COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, RECOGNITION PEPTIDE | | Authors: | Kanyo, Z.F, Pan, K.M, Williamson, R.A, Burton, D.R, Prusiner, S.B, Fletterick, R.J, Cohen, F.E. | | Deposit date: | 1999-08-20 | | Release date: | 2000-04-17 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change.
J.Mol.Biol., 293, 1999
|
|
2ZNN
 
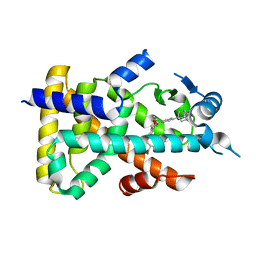 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist TIPP703 | | Descriptor: | (2S)-2-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Toyota, K, Kasuga, J, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZNQ
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP401 | | Descriptor: | (2S)-2-{3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]-4-methoxybenzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZNP
 
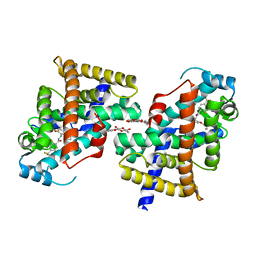 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP204 | | Descriptor: | (2S)-2-{4-butoxy-3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZNO
 
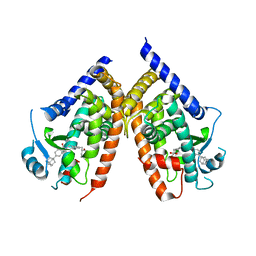 | | Human PPAR gamma ligand binding domain in complex with a synthetic agonist TIPP703 | | Descriptor: | (2S)-2-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Waku, T, Kasuga, J, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3L16
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3L17
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 4-methyl-5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyrimidin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
