8FHD
 
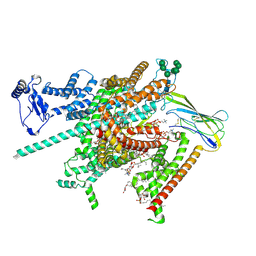 | | Cryo-EM structure of human voltage-gated sodium channel Nav1.6 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (5E,17R,20S)-23-amino-20-hydroxy-14,20-dioxo-15,19,21-trioxa-20lambda~5~-phosphatricos-5-en-17-yl hexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5NAJ
 
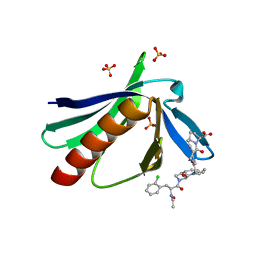 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-1]-[ProM-1]-OH | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{R},7~{S},10~{S},13~{R})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{S},7~{R},10~{R},13~{S})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, CHLORIDE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4GE8
 
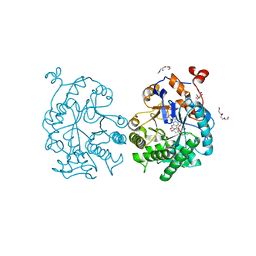 | | OYE1-W116I complexed with (s)-Carvone | | Descriptor: | (5S)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4B7D
 
 | |
4B7S
 
 | |
6ZXS
 
 | | Cold grown Pea Photosystem I | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Caspy, I, Borovikova-Sheinker, A, Subramanyam, R, Nelson, N. | | Deposit date: | 2020-07-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of cold grown pea Photosystem I
To Be Published
|
|
1XCX
 
 | | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Li, C, Begum, A, Numao, S, Park, K.H, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-03 | | Release date: | 2004-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts
Biochemistry, 44, 2005
|
|
1XCW
 
 | | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, ... | | Authors: | Li, C, Begum, A, Numao, S, Park, K.H, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-03 | | Release date: | 2004-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts
Biochemistry, 44, 2005
|
|
4FGG
 
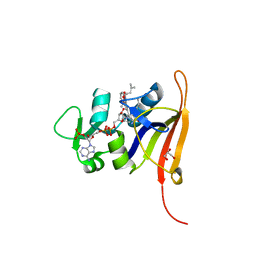 | | S. aureus dihydrofolate reductase co-crystallized with propyl-DAP isobutenyl-dihydrophthalazine inhibitor | | Descriptor: | (2E)-3-{5-[(2,4-diamino-6-propylpyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(2-methylprop-1-en-1-yl)phthalazin-2(1H)-yl]prop-2-en-1-one, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of Bacterial Dihydrofolate Reductase by 6-Alkyl-2,4-diaminopyrimidines.
Chemmedchem, 7, 2012
|
|
4H6B
 
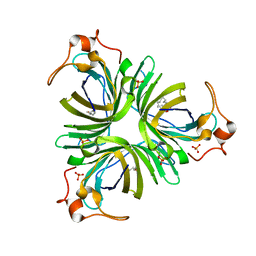 | | Structural basis for allene oxide cyclization in moss | | Descriptor: | (9Z)-11-{(2R,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, (9Z)-11-{(2S,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, Allene oxide cyclase, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of Physcomitrella patens AOC1 and AOC2: Insights into the Enzyme Mechanism and Differences in Substrate Specificity.
Plant Physiol., 160, 2012
|
|
4ELF
 
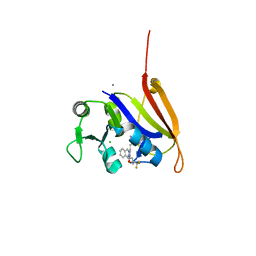 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(3,3,3-trifluoropropyl)phthalazin-2(1H)-yl ]prop-2-en-1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4GWE
 
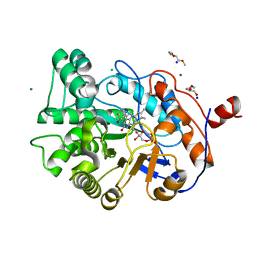 | | W116L-OYE1 complexed with (R)-carvone | | Descriptor: | (5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2012-09-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4BFD
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | BETA-SECRETASE 1, DIMETHYL SULFOXIDE, N-[3-[(1S,3S,6S)-5-azanyl-3-methyl-4-azabicyclo[4.1.0]hept-4-en-3-yl]-4-fluoranyl-phenyl]-5-chloranyl-pyridine-2-carbox amide, ... | | Authors: | Banner, D.W, Benz, J, Stihle, M. | | Deposit date: | 2013-03-18 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bace1 Inhibitors: A Head Group Scan on a Series of Amides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ELE
 
 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(propan-2-yl)phthalazin-2(1H)-yl]prop-2-en -1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
1B6A
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH TNP-470 | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
3M08
 
 | | Wild Type Dihydrofolate Reductase from Staphylococcus aureus with inhibitor RAB1 | | Descriptor: | 5-(3,4-dimethoxy-5-{(1E)-3-oxo-3-[(1S)-1-propylphthalazin-2(1H)-yl]prop-1-en-1-yl}benzyl)pyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.014 Å) | | Cite: | Inhibition of Antibiotic-Resistant Staphylococcus aureus by the Broad-Spectrum Dihydrofolate Reductase Inhibitor RAB1.
Antimicrob.Agents Chemother., 54, 2010
|
|
1B12
 
 | | CRYSTAL STRUCTURE OF TYPE 1 SIGNAL PEPTIDASE FROM ESCHERICHIA COLI IN COMPLEX WITH A BETA-LACTAM INHIBITOR | | Descriptor: | PHOSPHATE ION, SIGNAL PEPTIDASE I, prop-2-en-1-yl (2S)-2-[(2S,3R)-3-(acetyloxy)-1-oxobutan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylate | | Authors: | Paetzel, M, Dalbey, R, Strynadka, N.C.J. | | Deposit date: | 1999-11-24 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor.
Nature, 396, 1998
|
|
3M09
 
 | | F98Y TMP-resistant Dihydrofolate Reductase from Staphylococcus aureus with inhibitor RAB1 | | Descriptor: | 5-(3,4-dimethoxy-5-{(1E)-3-oxo-3-[(1S)-1-propylphthalazin-2(1H)-yl]prop-1-en-1-yl}benzyl)pyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Inhibition of Antibiotic-Resistant Staphylococcus aureus by the Broad-Spectrum Dihydrofolate Reductase Inhibitor RAB1.
Antimicrob.Agents Chemother., 54, 2010
|
|
2R1X
 
 | | Crystal structure of S25-2 Fab in complex with Kdo analogues | | Descriptor: | 2,6-anhydro-3,5-dideoxy-D-ribo-oct-2-enonic acid-(4-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Fab, antibody fragment (IgG1k), ... | | Authors: | Brooks, C.L, Evans, S.V. | | Deposit date: | 2007-08-23 | | Release date: | 2008-12-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes.
J.Mol.Biol., 377, 2008
|
|
1MXD
 
 | | Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
4WEJ
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | Descriptor: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4X6F
 
 | | CD1a binary complex with sphingomyelin | | Descriptor: | (4S,7S,23Z)-4-hydroxy-7-[(1S,2Z)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-9-oxo-3,5-dioxa-8-aza-4-phosphadotriacont- 23-en-1-aminium 4-oxide, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
7W9K
 
 | | Cryo-EM structure of human Nav1.7-beta1-beta2 complex at 2.2 angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
7PI5
 
 | | Unstacked stretched Dunaliella PSII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Fadeeva, M, Mazor, Y, Nelson, N. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structure of Dunaliella Photosystem II reveals conformational flexibility of stacked and unstacked supercomplexes.
Elife, 12, 2023
|
|
