2CPL
 
 | |
4B6S
 
 | | Structure of Helicobacter pylori Type II Dehydroquinase inhibited by (2S)-2-Perfluorobenzyl-3-dehydroquinic acid | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, PHOSPHATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lence, E, Tizon, L, Peon, A, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic basis of the inhibition of type II dehydroquinase by (2S)- and (2R)-2-benzyl-3-dehydroquinic acids.
ACS Chem. Biol., 8, 2013
|
|
4R59
 
 | |
3ESB
 
 | | cut-1c; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), CHLORIDE ION, Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
2E5W
 
 | |
5REL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102340 | | Descriptor: | 1-{4-[(3-methylphenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4JBG
 
 | | 1.75A resolution structure of a thermostable alcohol dehydrogenase from Pyrobaculum aerophilum | | Descriptor: | Alcohol dehydrogenase (Zinc), CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Vitale, A, Throne, N, Hu, X, Shen, M, D'Auria, S, Auld, D.S. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Physicochemical Characterization of a Thermostable Alcohol Dehydrogenase from Pyrobaculum aerophilum.
Plos One, 8, 2013
|
|
3ET3
 
 | |
2EBL
 
 | | Solution structure of the Zinc finger, C4-type domain of human COUP transcription factor 1 | | Descriptor: | COUP transcription factor 1, ZINC ION | | Authors: | Yoneyama, M, Koshiba, S, Watabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-09 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Zinc finger, C4-type domain of human COUP transcription factor 1
To be Published
|
|
2GAR
 
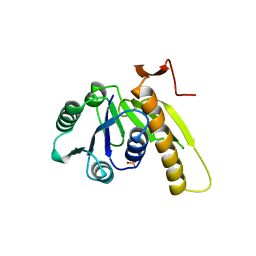 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
2ECM
 
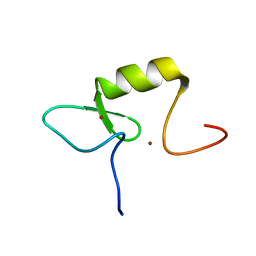 | | Solution structure of the RING domain of the RING finger and CHY zinc finger domain-containing protein 1 from Mus musculus | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, ZINC ION | | Authors: | Miyamoto, K, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-13 | | Release date: | 2007-08-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RING domain of the RING finger and CHY zinc finger domain-containing protein 1 from Mus musculus
To be Published
|
|
3R2N
 
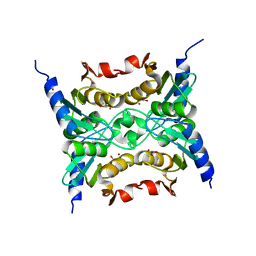 | |
2GAT
 
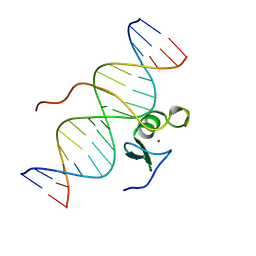 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
3R9T
 
 | |
3QD5
 
 | |
1DOS
 
 | | STRUCTURE OF FRUCTOSE-BISPHOSPHATE ALDOLASE | | Descriptor: | ALDOLASE CLASS II, AMMONIUM ION, ZINC ION | | Authors: | Blom, N, Tetreault, S, Coulombe, R, Sygusch, J. | | Deposit date: | 1996-06-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 3, 1996
|
|
4RLO
 
 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
3QQS
 
 | | Anthranilate phosphoribosyltransferase (TRPD) from Mycobacterium tuberculosis (complex with inhibitor ACS172) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2,2'-iminodibenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Short, F.L, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-02-16 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Substrate Capture Mechanism of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Provides a Mode for Inhibition.
Biochemistry, 52, 2013
|
|
1XDF
 
 | | Crystal structure of pathogenesis-related protein LlPR-10.2A from yellow lupine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PR10.2A, SODIUM ION | | Authors: | Pasternak, O, Biesiadka, J, Dolot, R, Handschuh, L, Bujacz, G, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2004-09-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a yellow lupin pathogenesis-related PR-10 protein belonging to a novel subclass.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3F7I
 
 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2008-11-09 | | Release date: | 2009-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orally bioavailable antagonists of inhibitor of apoptosis proteins based on an azabicyclooctane scaffold
J.Med.Chem., 52, 2009
|
|
2E5K
 
 | | Solution structure of SH3 domain in Suppressor of T-cell receptor signaling 1 | | Descriptor: | Suppressor of T-cell receptor signaling 1 | | Authors: | Sukegawa, S, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-21 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3 domain in Suppressor of T-cell receptor signaling 1
To be Published
|
|
2K3S
 
 | | HADDOCK-derived structure of the CH-domain of the smoothelin-like 1 complexed with the C-domain of apocalmodulin | | Descriptor: | Calmodulin, Smoothelin-like protein 1 | | Authors: | Ishida, H, Borman, M.A, Ostrander, J, Vogel, H.J, MacDonald, J.A. | | Deposit date: | 2008-05-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calponin homology (CH) domain from the smoothelin-like 1 protein: a unique apocalmodulin-binding mode and the possible role of the C-terminal type-2 CH-domain in smooth muscle relaxation.
J.Biol.Chem., 283, 2008
|
|
4IMF
 
 | |
3V49
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 1 | | Descriptor: | 4-[(4R)-4-(4-hydroxyphenyl)-3,4-dimethyl-2,5-dioxoimidazolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, activator peptide, ... | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.-M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
4RH2
 
 | |
