5LDW
 
 | | Structure of mammalian respiratory Complex I, class1 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
7ENA
 
 | | TFIID-based PIC-Mediator holo-complex in pre-assembled state (pre-hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7DWQ
 
 | | Photosystem I from a chlorophyll d-containing cyanobacterium Acaryochloris marina | | Descriptor: | (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Chen, J.H, Zhang, X, Shen, J.R. | | Deposit date: | 2021-01-17 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina.
J Integr Plant Biol, 63, 2021
|
|
8B64
 
 | | Cryo-EM structure of RC-LH1-PufX photosynthetic core complex from Rba. capsulatus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.589 Å) | | Cite: | Cryo-EM structure of a monomeric RC-LH1-PufX supercomplex with high-carotenoid content from Rhodobacter capsulatus.
Structure, 31, 2023
|
|
7ENC
 
 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
8BIP
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
6VPV
 
 | | Trimeric Photosystem I from the High-Light Tolerant Cyanobacteria Cyanobacterium Aponinum | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Dobson, Z, Toporik, H, Vaughn, N, Lin, S, Williams, D, Fromme, P, Mazor, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structure of photosystem I from a high-light tolerant Cyanobacteria.
Elife, 10, 2021
|
|
7ZUW
 
 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZPQ
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C1) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZMK
 
 | |
8UQL
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UQP
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URH
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 27 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
8UR0
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
6X2I
 
 | |
7EJ4
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb-25, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-25
To be published
|
|
7EG2
 
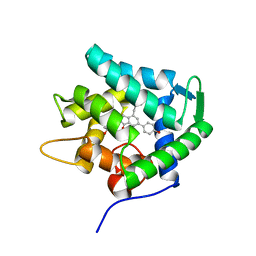 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
6WDF
 
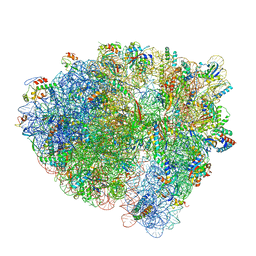 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure VI-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
7EB0
 
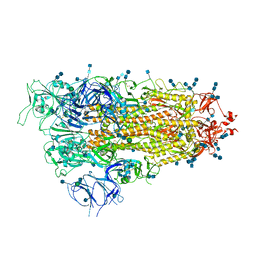 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EAZ
 
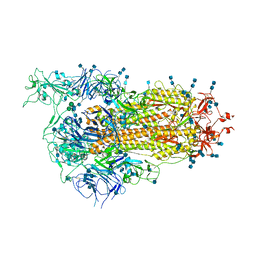 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
6WDE
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure V-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
8UEQ
 
 | | In-situ complex I with Q10 (State-beta) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
7EJ5
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb45, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody RBD-chAb-45
To Be Published
|
|
8UEP
 
 | | In-situ complex I, Active-Q10 (State-alpha) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
