5SVD
 
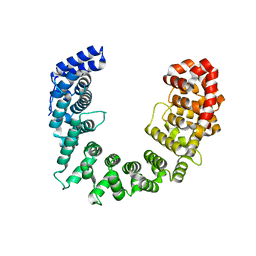 | |
4GCW
 
 | | Crystal structure of RNase Z in complex with precursor tRNA(Thr) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ribonuclease Z, TRNA(THR), ... | | Authors: | Pellegrini, O, Li de la Sierra-Gallay, I, Piton, J, Gilet, L, Condon, C. | | Deposit date: | 2012-07-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation of tRNA Maturation by Downstream Uracil Residues in B. subtilis
Structure, 20, 2012
|
|
3TWH
 
 | | Selenium Derivatized RNA/DNA Hybrid in complex with RNase H Catalytic Domain D132N Mutant | | Descriptor: | DNA (5'-D(*AP*TP*(SDG)P*TP*CP*(SDG))-3'), MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rob, A, Gerlits, O, Jiang, J.S, Gan, J.H, Huang, Z. | | Deposit date: | 2011-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Novel complex MAD phasing and RNase H structural insights using selenium oligonucleotides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
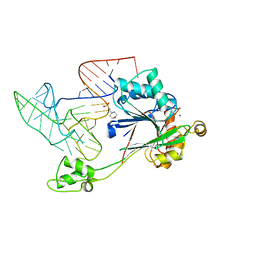
2D0C
 
 | | Crystal structure of Bst-RNase HIII in complex with Mn2+ | | Descriptor: | MANGANESE (II) ION, ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
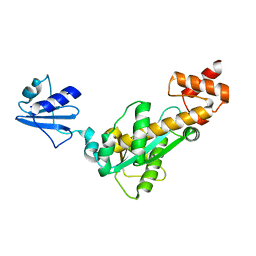
2D0B
 
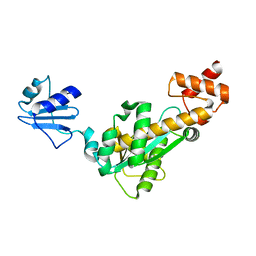 | | Crystal structure of Bst-RNase HIII in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
2D0A
 
 | | Crystal structure of Bst-RNase HIII | | Descriptor: | ribonuclease HIII | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-07-31 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus
J.Mol.Biol., 356, 2006
|
|
3DKP
 
 | | Human DEAD-box RNA-helicase DDX52, conserved domain I in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lehtio, L, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Wisniewska, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2IWD
 
 | | Oxacilloyl-acylated MecR1 extracellular antibiotic-sensor domain. | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Methicillin resistance mecR1 protein | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
5N8M
 
 | |
5N8L
 
 | |
8WFB
 
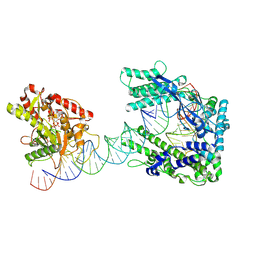 | | Cryo-EM structure of the dPspCas13b-ADAR2-crRNA-target RNA complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, ZINC ION, crRNA, ... | | Authors: | Ishikawa, J, Kato, K, Yamashita, K, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2023-09-19 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into RNA-guided RNA editing by the Cas13b-ADAR2 complex.
Nat.Struct.Mol.Biol., 2025
|
|
5BZU
 
 | |
8PUW
 
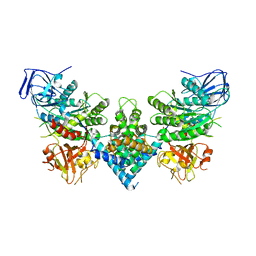 | | Chaetomium thermophilum Las1-Grc3-complex | | Descriptor: | Las1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
7F36
 
 | | TacT complexed with acetyl-glycyl-tRNAGly | | Descriptor: | ACETYLAMINO-ACETIC ACID, CALCIUM ION, N-acetyltransferase domain-containing protein, ... | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Molecular basis of glycyl-tRNAGly acetylation by TacT from Salmonella Typhimurium
Cell Rep, 37, 2021
|
|
1I5L
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH SHORT POLY-U RNA | | Descriptor: | 5'-R(*UP*UP*U)-3', PUTATIVE SNRNP SM-LIKE PROTEIN AF-SM1, URIDINE | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-28 | | Release date: | 2001-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
4XHA
 
 | | Crystal structure of Thosea asigna virus RNA-dependent RNA polymerase (RdRP) complexed with Lu3+ | | Descriptor: | GLYCEROL, LUTETIUM (III) ION, RNA-dependent RNA polymerase, ... | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-01-05 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
1MWH
 
 | | REOVIRUS POLYMERASE LAMBDA3 BOUND TO MRNA CAP ANALOG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MANGANESE (II) ION, MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
4XHI
 
 | | Crystal structure of native Thosea asigna virus RNA-dependent RNA polymerase (RdRP) at 2.15 Angstrom resolution | | Descriptor: | GLYCEROL, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-01-05 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
7TNX
 
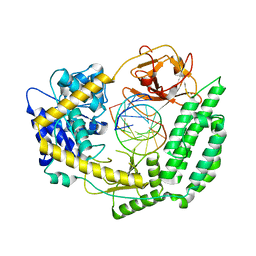 | | Cryo-EM structure of RIG-I in complex with p3dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3dsRNAa, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
5I62
 
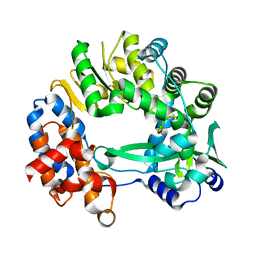 | |
7JNH
 
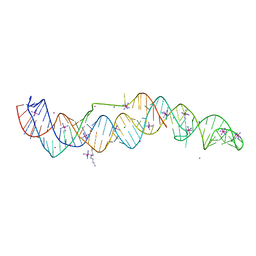 | | Crystal structure of a double-ENE RNA stability element in complex with a 28-mer poly(A) RNA | | Descriptor: | 28-mer poly(A) RNA, COBALT HEXAMMINE(III), Core double ENE RNA (Xtal construct) from Oryza sativa transposon,Core double ENE RNA (Xtal construct) from Oryza sativa transposon, ... | | Authors: | Torabi, S.F, Vaidya, A.T, Tycowski, K.T, DeGregorio, S.J, Wang, J, Shu, M.D, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | RNA stabilization by a poly(A) tail 3'-end binding pocket and other modes of poly(A)-RNA interaction.
Science, 371, 2021
|
|
1MP1
 
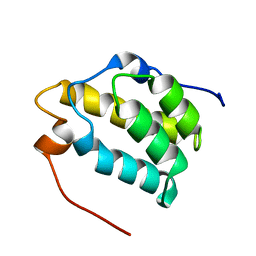 | | Solution structure of the PWI motif from SRm160 | | Descriptor: | Ser/Arg-related nuclear matrix protein | | Authors: | Szymczyna, B.R, Bowman, J, McCracken, S, Pineda-Lucena, A, Lu, Y, Cox, B, Lambermon, M, Graveley, B.R, Arrowsmith, C.H, Blencowe, B.J. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing.
Genes Dev., 17, 2003
|
|
5MSF
 
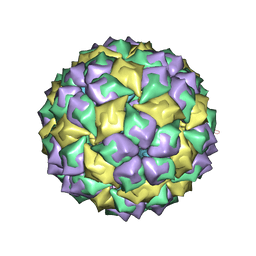 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*CP*CP*GP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-15 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
6TME
 
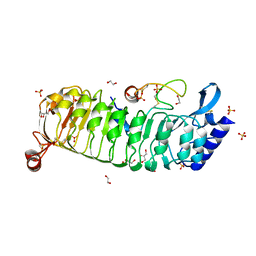 | | Monomeric LRX8 in complex with RALF4. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(3-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moussu, S, Caroline, C, Santos-Fernandez, G, Wehrle, S, Grossniklaus, U, Santiago, J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TJV
 
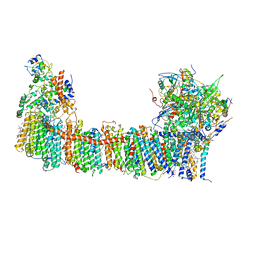 | | Structure of the NDH-1MS complex from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, BETA-CAROTENE, ... | | Authors: | Schuller, J.M, Saura, P, Thiemann, J, Schuller, S.K, Gamiz-Hernandez, A.P, Kurisu, G, Nowaczyk, M.M, Kaila, V.R.I. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I.
Nat Commun, 11, 2020
|
|
