6UDK
 
 | | HIV-1 bNAb 1-55 in complex with modified BG505 SOSIP-based immunogen RC1 and 10-1074 | | Descriptor: | 1-55 Fab Heavy Chain, 1-55 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
7YZQ
 
 | | MgADP-AlF4-bound DCCP:DCCP-R complex | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, AMMONIUM ION, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for coupled ATP-driven electron transfer in the double-cubane cluster protein.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YZM
 
 | | MgADPNP-bound DCCP:DCCP-R complex | | Descriptor: | (R,R)-2,3-BUTANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for coupled ATP-driven electron transfer in the double-cubane cluster protein.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z4B
 
 | | Bacteriophage SU10 virion (C1) | | Descriptor: | Adaptor, Major head protein, Portal protein, ... | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7BOF
 
 | | Bacterial 30S ribosomal subunit assembly complex state I (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8U7Q
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
6WPS
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
7BOD
 
 | | Bacterial 30S ribosomal subunit assembly complex state M (body domain) | | Descriptor: | 16S rRNA (body domain of 30S subunit), 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
6U4P
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-26 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
5J5F
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with N4,N4-bis[(pyridin-2-yl)methyl]-6-(thiophen-3-yl)pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczanowska, K, Camacho Hernandez, G.A, Harel, M, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
8UD6
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
7YQ1
 
 | | Crystal structure of adenosine 5'-phosphosulfate kinase from Archaeoglobus fulgidus in complex with AMP-PNP and APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Adenylyl-sulfate kinase, BORIC ACID, ... | | Authors: | Kawakami, T, Teramoto, T, Kakuta, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of adenosine 5'-phosphosulfate kinase isolated from Archaeoglobus fulgidus.
Biochem.Biophys.Res.Commun., 643, 2022
|
|
7BMH
 
 | | Crystal structure of a light-driven proton pump LR (Mac) from Leptosphaeria maculans | | Descriptor: | EICOSANE, OLEIC ACID, Opsin | | Authors: | Kovalev, K, Zabelskii, D, Dmitrieva, N, Volkov, O, Shevchenko, V, Astashkin, R, Zinovev, E, Gordeliy, V. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based insights into evolution of rhodopsins.
Commun Biol, 4, 2021
|
|
7BOG
 
 | | Bacterial 30S ribosomal subunit assembly complex state E (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
5JIA
 
 | |
8UFU
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(9-amino-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)-4-methylquinolin-2-amine | | Descriptor: | (7M)-7-[(9S)-9-amino-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl]-4-methylquinolin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
5JJM
 
 | |
8UK2
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 5 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
8UK3
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 6 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
6U9V
 
 | |
8UPP
 
 | | Campylobacter jejuni ketol-acid reductoisomerase in complex with NADPH and Hoe704 | | Descriptor: | (2R)-(dimethylphosphoryl)(hydroxy)acetic acid, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Lin, X, Lv, Y, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2023-10-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mapping of the Reaction Trajectory catalyzed by Class I Ketol-Acid Reductoisomerase
Acs Catalysis, 2024
|
|
5JKM
 
 | |
6U7H
 
 | | Cryo-EM structure of the HCoV-229E spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Z, Benlekbir, S, Rubinstein, J.L, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6WP0
 
 | |
5JMN
 
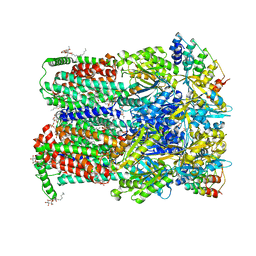 | | Fusidic acid bound AcrB | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, DARPin, ... | | Authors: | Oswald, C, Tam, H.K, Pos, K.M. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB.
Nat Commun, 7, 2016
|
|
