4PS5
 
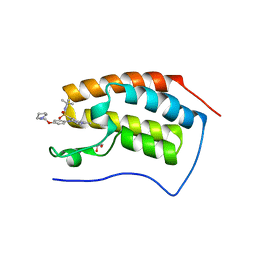 | | Crystal structure of the first bromodomain of human BRD4 in complex with TG101348 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bromodomain-containing protein 4, ... | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2014-03-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
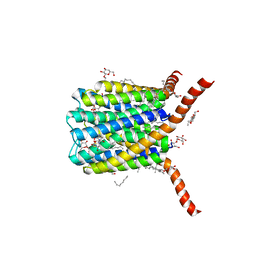
5ZE3
 
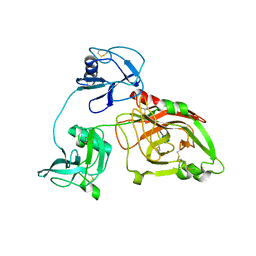 | |
2PNO
 
 | | Crystal structure of human leukotriene C4 synthase | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, Leukotriene C4 synthase | | Authors: | Ago, H, Kanaoka, Y, Irikura, D, Lam, B.K, Shimamura, T, Austen, K.F, Miyano, M. | | Deposit date: | 2007-04-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis
Nature, 448, 2007
|
|
2X89
 
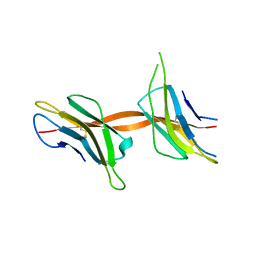 | | Structure of the Beta2_microglobulin involved in amyloidogenesis | | Descriptor: | ANTIBODY, BETA-2-MICROGLOBULIN | | Authors: | Domanska, K, Srinivasan, V, Vanderhaegen, S, Pardon, E, Marquez, J.A, Bellotti, V, Wyns, L, Steyaert, J. | | Deposit date: | 2010-03-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Atomic Structure of a Nanobody-Trapped Domain-Swapped Dimer of an Amyloidogenic {Beta}2-Microglobulin Variant.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5ZE9
 
 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
2XAC
 
 | |
5DQN
 
 | | Polyethylene 600-bound form of P450 CYP125A3 mutant from Myobacterium Smegmatis - W83Y | | Descriptor: | CITRIC ACID, Cytochrome P450 CYP125, PENTAETHYLENE GLYCOL, ... | | Authors: | Ortiz de Montellano, P.J, Frank, D.J, Waddling, C.A. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Cytochrome P450 125A4, the Third Cholesterol C-26 Hydroxylase from Mycobacterium smegmatis.
Biochemistry, 54, 2015
|
|
5A4N
 
 | |
4PSV
 
 | | Mycobacterium tuberculosis RecA phosphate bound room temperature structure I-RT | | Descriptor: | PHOSPHATE ION, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-08 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
5DR0
 
 | | Endothiapepsin in complex with fragment 203 | | Descriptor: | 1,2-ETHANEDIOL, 5-(methylsulfanyl)-4-(propan-2-ylsulfonyl)-1H-pyrazol-3-amine, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4PT7
 
 | | Structure of initiator | | Descriptor: | Replication initiator A family protein, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CBB
 
 | | Crystal Structure of Hepatocyte Nuclear Factor 4alpha in complex with DNA: Diabetes Gene Product | | Descriptor: | Hepatocyte Nuclear Factor 4-alpha promoter element DNA, Hepatocyte Nuclear Factor 4-alpha, DNA binding domain, ... | | Authors: | Lu, P, Rha, G.B, Melikishvili, M, Adkins, B.C, Fried, M.G, Chi, Y.I. | | Deposit date: | 2008-02-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of natural promoter recognition by a unique nuclear receptor, HNF4alpha. Diabetes gene product.
J.Biol.Chem., 283, 2008
|
|
4IEF
 
 | | Complex of Porphyromonas gingivalis RgpB pro- and mature domains | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BARIUM ION, CALCIUM ION, ... | | Authors: | de Diego, I, Veillard, F.T, Guevara, T, Potempa, B, Sztukowska, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porphyromonas gingivalis Virulence Factor Gingipain RgpB Shows a Unique Zymogenic Mechanism for Cysteine Peptidases.
J.Biol.Chem., 288, 2013
|
|
3G5H
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5G6M
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 7-((3-Aminomethyl)phenoxy)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[3-(aminomethyl)phenoxy]methyl]quinolin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2016-06-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors.
Biochemistry, 55, 2016
|
|
5DRG
 
 | | Green/cyan WasCFP at pH 10.0 | | Descriptor: | GLYCEROL, Green/cyan WasCFP_pH10 at pH 10.0, SODIUM ION | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
4ZTP
 
 | |
4ZTX
 
 | | Neurospora crassa cobalamin-independent methionine synthase complexed with Zn2+ | | Descriptor: | Cobalamin-Independent Methionine synthase, GLYCEROL, NITRATE ION, ... | | Authors: | Wheatley, R.W, Ng, K.K, Kapoor, M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal cobalamin-independent methionine synthase: Insights from the model organism, Neurospora crassa.
Arch.Biochem.Biophys., 590, 2015
|
|
4PUJ
 
 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 6-Amino-2-{[2-(morpholin-4-yl)ethyl]amino}-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-[(2-morpholin-4-ylethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chasing Protons: How Isothermal Titration Calorimetry, Mutagenesis, and pKa Calculations Trace the Locus of Charge in Ligand Binding to a tRNA-Binding Enzyme.
J.Med.Chem., 57, 2014
|
|
4ZYB
 
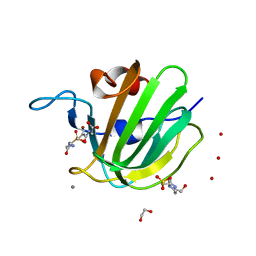 | | High resolution structure of M23 peptidase LytM with substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Grabowska, M, Jagielska, E, Czapinska, H, Bochtler, M, Sabala, I. | | Deposit date: | 2015-05-21 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of an M23 peptidase with a substrate analogue.
Sci Rep, 5, 2015
|
|
5ZF0
 
 | | X-ray Structure of the Electron Transfer Complex between Ferredoxin and Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kubota-Kawai, H, Mutoh, R, Shinmura, K, Setif, P, Nowaczyk, M, Roegner, M, Ikegami, T, Tanaka, T, Kurisu, G. | | Deposit date: | 2018-03-01 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | X-ray structure of an asymmetrical trimeric ferredoxin-photosystem I complex
Nat Plants, 4, 2018
|
|
3CBT
 
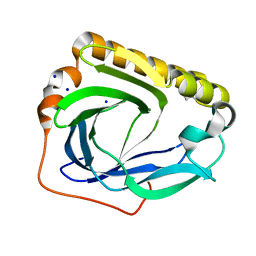 | | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor | | Descriptor: | MAGNESIUM ION, Phosphatase SC4828, SODIUM ION | | Authors: | Singer, A.U, Xu, X, Chang, C, Zheng, H, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor.
To be Published
|
|
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
5ZJ7
 
 | |
4PCH
 
 | |
