4E01
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with AMPPNP | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
4E00
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with ADP | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with ADP
To be Published
|
|
4H81
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-2-chloro-3-phenylpropanoic acid complex with ADP | | Descriptor: | (2R)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-09-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4XIV
 
 | |
6SWJ
 
 | | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus (with Pt) | | Descriptor: | Histidine kinase, PLATINUM (II) ION | | Authors: | Lansky, S, Shiradski, M, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus (with Pt)
To Be Published
|
|
6SWK
 
 | | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus | | Descriptor: | Histidine kinase | | Authors: | Lansky, S, Shiradsky, M, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus
To Be Published
|
|
2ZDY
 
 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-12-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZDX
 
 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Pyruvate dehydrogenase kinase isozyme 4 | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZBK
 
 | | Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms | | Descriptor: | RADICICOL, Type 2 DNA topoisomerase 6 subunit B, Type II DNA topoisomerase VI subunit A | | Authors: | Graille, M, Cladiere, L, Durand, D, Lecointe, F, Forterre, P, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2007-10-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Crystal Structure of an Intact Type II DNA Topoisomerase: Insights into DNA Transfer Mechanisms
Structure, 16, 2008
|
|
2ZKJ
 
 | | Crystal structure of human PDK4-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Li, J, Chuang, D.T. | | Deposit date: | 2008-03-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyruvate Dehydrogenase Kinase-4 Structures Reveal a Metastable Open Conformation Fostering Robust Core-free Basal Activity
J.Biol.Chem., 283, 2008
|
|
1Z5C
 
 | | Topoisomerase VI-B, ADP Pi bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
2BU8
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DICHLORO-ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2BU2
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2BU7
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2BTZ
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2BU6
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | (N-{4-[(ETHYLANILINO)SULFONYL]-2-METHYLPHENYL}-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
1Z5B
 
 | |
2BU5
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
1Z5A
 
 | | Topoisomerase VI-B, ADP-bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1Z59
 
 | | Topoisomerase VI-B, ADP-bound monomer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
7EBG
 
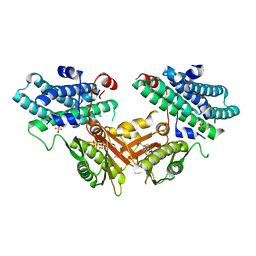 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 7 | | Descriptor: | 3,3-dimethyl-7-(methylamino)-1H-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EAT
 
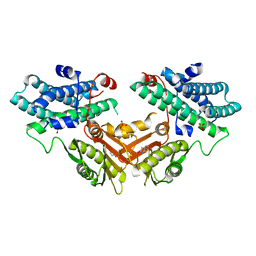 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 1 | | Descriptor: | 1,3-dihydro-2H-indol-2-one, SULFATE ION, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBB
 
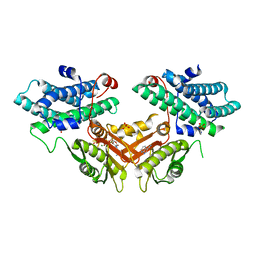 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 2 | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EA0
 
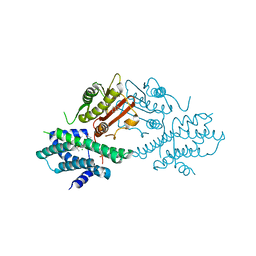 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 1 | | Descriptor: | 1,3-dihydro-2H-indol-2-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-05 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBH
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 13 | | Descriptor: | 5-bromanyl-2-methyl-6-propyl-7H-pyrrolo[2,3-d]pyrimidine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
