7Y5U
 
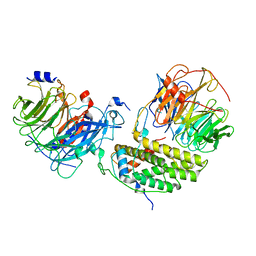 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
1VYB
 
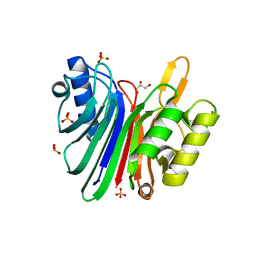 | | Endonuclease domain of human LINE1 ORF2p | | Descriptor: | GLYCEROL, ORF2 CONTAINS A REVERSE TRANSCRIPTASE DOMAIN, SULFATE ION, ... | | Authors: | Weichenrieder, O, Repanas, K, Perrakis, A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-06-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon.
Structure, 12, 2004
|
|
8BRN
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with an alpha-aminonaphthylmethylphosphonic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Hussein, W.M, Guddat, L.W, Schenk, G. | | Deposit date: | 2022-11-23 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of an alpha-aminonaphthylmethylphosphonic acid inhibitor of purple acid phosphatase using rational structure-based design approaches.
Eur.J.Med.Chem., 254, 2023
|
|
6ROH
 
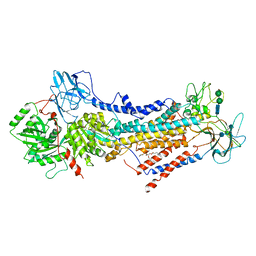 | | Cryo-EM structure of the autoinhibited Drs2p-Cdc50p | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
3UGD
 
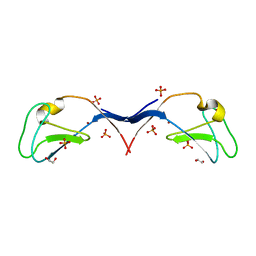 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Protein kinase C delta type, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta
Biophys.J., 103, 2012
|
|
1Z30
 
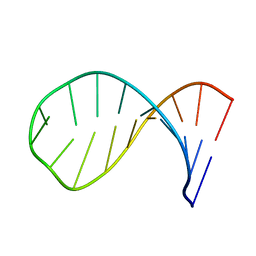 | | NMR structure of the apical part of stemloop D from cloverleaf 1 of bovine enterovirus 1 RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*CP*GP*UP*UP*AP*GP*AP*AP*CP*GP*UP*C)-3' | | Authors: | Ihle, Y, Ohlenschlager, O, Duchardt, E, Ramachandran, R, Gorlach, M. | | Deposit date: | 2005-03-10 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA
Nucleic Acids Res., 33, 2005
|
|
1AKE
 
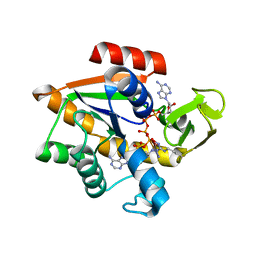 | |
3M7K
 
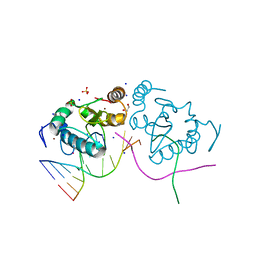 | | Crystal structure of PacI-DNA Enzyme product complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*AP*AP*T)-3'), DNA (5'-D(P*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
4LCT
 
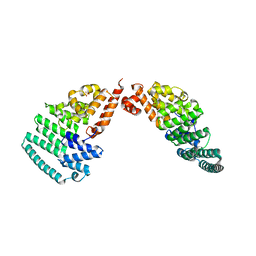 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6M4L
 
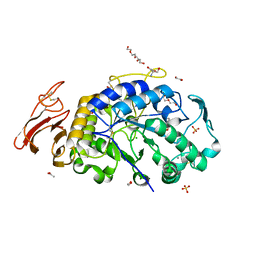 | | X-ray crystal structure of the E249Q mutant of alpha-amylase I from Eisenia fetida | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2NTT
 
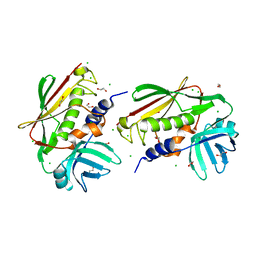 | | Crystal Structure of SEK | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Staphylococcal enterotoxin K | | Authors: | Gunther, S, Varma, A.K, Moza, B, Sundberg, E.J. | | Deposit date: | 2006-11-08 | | Release date: | 2007-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | A novel loop domain in superantigens extends their T cell receptor recognition site
J.Mol.Biol., 371, 2007
|
|
1W32
 
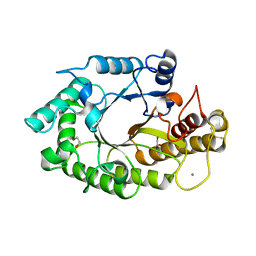 | | The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Andrews, S, Taylor, E.J, Pell, G.N, Vincent, F, Ducros, V.M.A, Davies, G.J, Lakey, J.H, Gilbert, H.J. | | Deposit date: | 2004-07-12 | | Release date: | 2004-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
2XCS
 
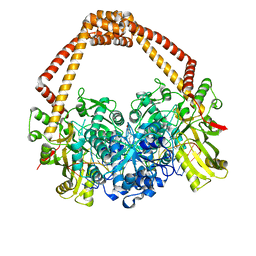 | | The 2.1A crystal structure of S. aureus Gyrase complex with GSK299423 and DNA | | Descriptor: | 5'-5UA*D(GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP *CP*TP*AP*CP*GP*GP*CP*T)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, DNA GYRASE SUBUNIT B, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
3AS1
 
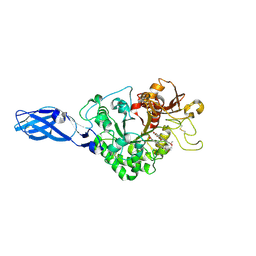 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with chelerythrine | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
1W9L
 
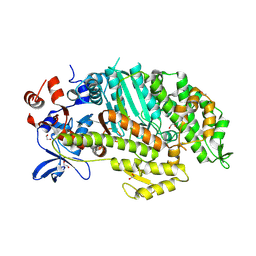 | | Myosin II Dictyostelium discoideum motor domain S456E bound with MgADP-AlF4 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
4LM8
 
 | |
6CQY
 
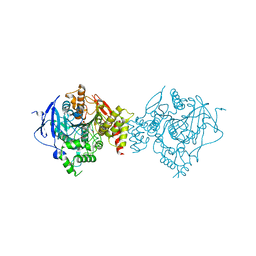 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with EMPA and HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Bester, S.M, Guelta, M.A, Pegan, S.D, Height, J.J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structural Insights of Stereospecific Inhibition of Human Acetylcholinesterase by VX and Subsequent Reactivation by HI-6.
Chem. Res. Toxicol., 31, 2018
|
|
6RV8
 
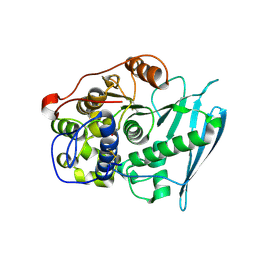 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
1LOL
 
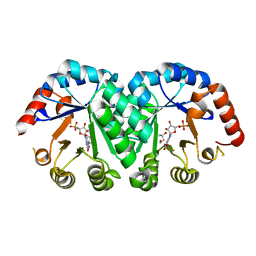 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
3M3J
 
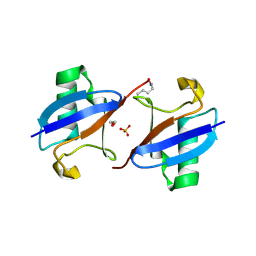 | | A new crystal form of Lys48-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Ubiquitin | | Authors: | Trempe, J.F, Brown, N.R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new crystal form of Lys48-linked diubiquitin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7FY8
 
 | | Crystal Structure of human FABP1 in complex with 2-[[4-cyclopropyl-5-methyl-3-[3-(trifluoromethyl)-1,2,4-oxadiazol-5-yl]thiophen-2-yl]carbamoyl]cyclopentene-1-carboxylic acid | | Descriptor: | 2-({(3P)-4-cyclopropyl-5-methyl-3-[3-(trifluoromethyl)-1,2,4-oxadiazol-5-yl]thiophen-2-yl}carbamoyl)cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, liver, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4CL1
 
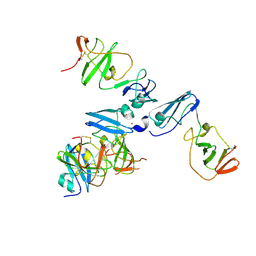 | | The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors | | Descriptor: | NON-STRUCTURAL PROTEIN 5A, SULFATE ION, ZINC ION | | Authors: | Lambert, S.M, Langley, D.R, Garnett, J.A, Angell, R, Hedgethorne, K, Meanwell, N.A, Matthews, S.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of Ns5A Domain 1 from Genotype 1A Reveals New Clues to the Mechanism of Action for Dimeric Hcv Inhibitors.
Protein Sci., 23, 2014
|
|
7G0E
 
 | | Crystal Structure of human FABP5 in complex with 2-[(3-ethoxycarbonyl-4,5,6,7-tetrahydro-1-benzothiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES C1CCC2=C(C1)C(=C(S2)NC(=O)C1=C(C(=O)O)CCC1)C(=O)OCC with IC50=1.1 microM | | Descriptor: | 2-{[3-(ethoxycarbonyl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4L8O
 
 | |
6LMQ
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase catalytic, SULFATE ION, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
