4QVC
 
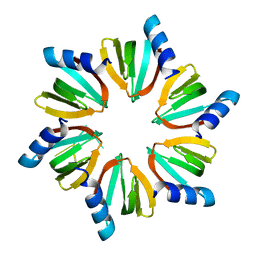 | | E.coli Hfq in complex with RNA Aus | | Descriptor: | RNA (5'-R(*AP*U*AP*AP*CP*UP*A)-3'), RNA-binding protein Hfq | | Authors: | Wang, L.J, Wang, W.W, Li, F.D, Wu, J.H, Gong, Q.G, Shi, Y.Y. | | Deposit date: | 2014-07-14 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq
Nucleic Acids Res., 43, 2015
|
|
4YE2
 
 | |
6ROL
 
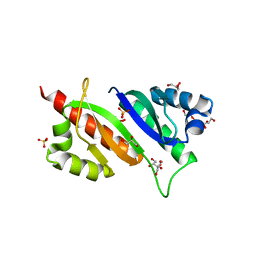 | | Structure of IMP2 KH34 domains | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 2, ... | | Authors: | Bhaskar, V, Biswas, J, Singer, R.H, Chao, J.A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for RNA selectivity by the IMP family of RNA-binding proteins.
Nat Commun, 10, 2019
|
|
7NXX
 
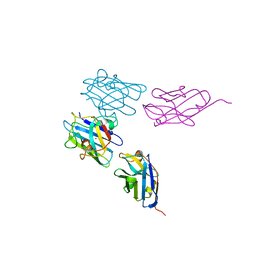 | | Structure of Superoxide Dismutase 1 (SOD1) in complex with nanobody 2 (Nb2). | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION, ... | | Authors: | Gallardo, R, Rousseau, F, Schymkowitz, J, Ulens, C. | | Deposit date: | 2021-03-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Identification and rational improvement of a nanobody that suppresses aggregation of mutant SOD1
To Be Published
|
|
5KDX
 
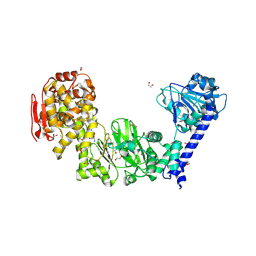 | | IMPa metallopeptidase in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, PHOSPHATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KD8
 
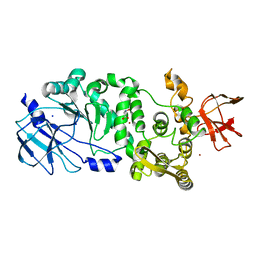 | | BT_4244 metallopeptidase in complex with Tn antigen. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Metallopeptidase, NICKEL (II) ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-07 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7NOZ
 
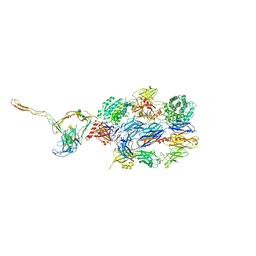 | | Structure of the nanobody stablized properdin bound alternative pathway proconvertase C3b:FB:FP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Lorenzen, J, Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2021-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Protein Sci., 31, 2022
|
|
6ZM2
 
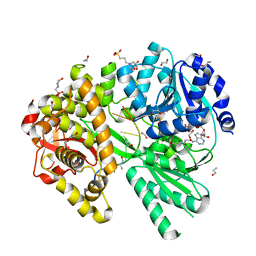 | |
5KDV
 
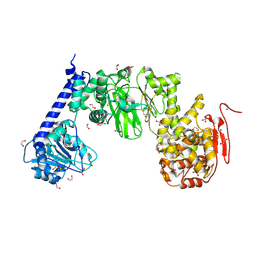 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, SULFATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6AVY
 
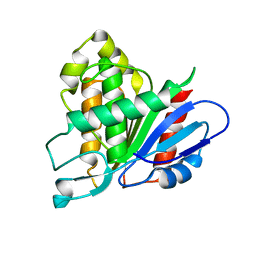 | |
7A9I
 
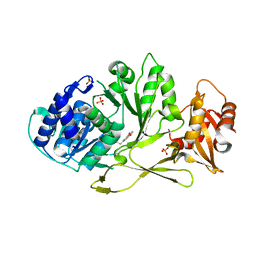 | |
7NMO
 
 | | Crystal structure of beta-2-microglobulin D76A mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMR
 
 | | Crystal structure of beta-2-microglobulin D76S mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6U5M
 
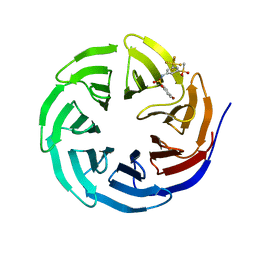 | |
6I50
 
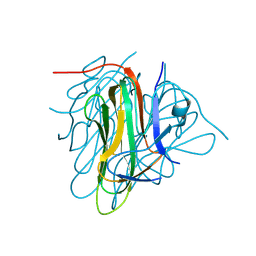 | |
7A9J
 
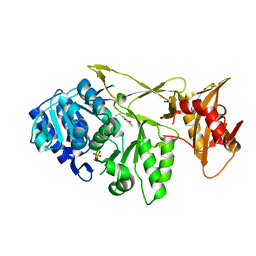 | |
6AR1
 
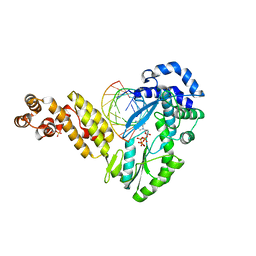 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Nat)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
7NMT
 
 | | Crystal structure of beta-2-microglobulin D76G mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMC
 
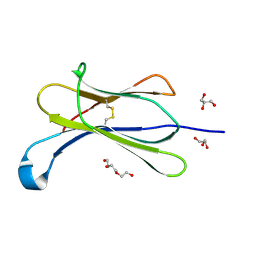 | | Crystal structure of beta-2-microglobulin D76E mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6U7F
 
 | | HCoV-229E RBD Class IV in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
3DSX
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with di-prenylated peptide Ser-Cys(GG)-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
7NMV
 
 | | Crystal structure of beta-2-microglobulin D76Q mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMY
 
 | | Crystal structure of beta-2-microglobulin D76Y mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6U8B
 
 | |
6U8P
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpA DNA | | Descriptor: | CpGpA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-05 | | Release date: | 2020-06-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
