1YLS
 
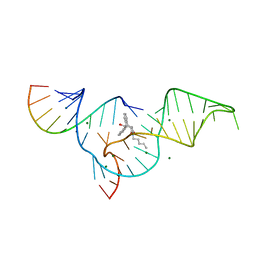 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YLT
 
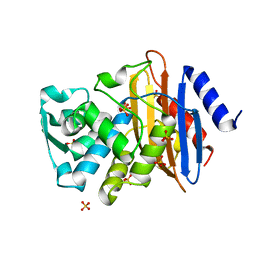 | | Atomic resolution structure of CTX-M-14 beta-lactamase | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-lactamase CTX-M-14 | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YLU
 
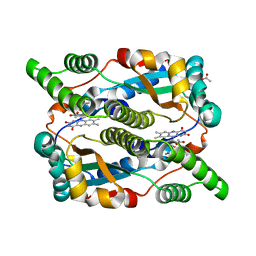 | | The structure of E. coli nitroreductase with bound acetate, crystal form 2 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1YLV
 
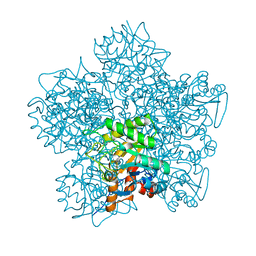 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID | | Descriptor: | LAEVULINIC ACID, PROTEIN (5-AMINOLAEVULINIC ACID DEHYDRATASE), ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Roper, J, Coker, A, Warren, M.J, Shoolingin-Jordan, P.M, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-02-22 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Schiff base complex of yeast 5-aminolaevulinic acid dehydratase with laevulinic acid.
Protein Sci., 8, 1999
|
|
1YLW
 
 | | X-ray structure of CTX-M-16 beta-lactamase | | Descriptor: | CTX-M-16 beta-lactamase, SULFATE ION | | Authors: | Chen, Y, Delmas, J, Sirot, J, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Atomic Resolution Structures of CTX-M beta-Lactamases: Extended Spectrum Activities from Increased Mobility and Decreased Stability.
J.Mol.Biol., 348, 2005
|
|
1YLX
 
 | |
1YLY
 
 | | X-ray crystallographic structure of CTX-M-9 beta-lactamase complexed with ceftazidime-like boronic acid | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase CTX-M-9 | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YLZ
 
 | | X-ray crystallographic structure of CTX-M-14 beta-lactamase complexed with ceftazidime-like boronic acid | | Descriptor: | BETA-LACTAMASE CTX-M-14, PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YM0
 
 | | Crystal Structure of Earthworm Fibrinolytic Enzyme Component B: a Novel, Glycosylated Two-chained Trypsin | | Descriptor: | MAGNESIUM ION, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, F, Wang, C, Li, M, Zhang, J.P, Gui, L.L, An, X.M, Chang, W.R. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of earthworm fibrinolytic enzyme component B: a novel, glycosylated two-chained trypsin.
J.Mol.Biol., 348, 2005
|
|
1YM1
 
 | | X-ray crystallographic structure of CTX-M-9 beta-lactamase complexed with a boronic acid inhibitor (SM2) | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, PHOSPHATE ION, beta-lactamase CTX-M-9a | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1YM2
 
 | | Crystal structure of human beta secretase complexed with NVP-AUR200 | | Descriptor: | Beta-secretase 1, NVP-AUR200 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
1YM3
 
 | | Crystal Structure of carbonic anhydrase RV3588c from Mycobacterium tuberculosis | | Descriptor: | CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), MAGNESIUM ION, ZINC ION | | Authors: | Covarrubias, A.S, Larsson, A.M, Hogbom, M, Lindberg, J, Bergfors, T, Bjorkelid, C, Mowbray, S.L, Unge, T, Jones, T.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and function of carbonic anhydrases from Mycobacterium tuberculosis.
J.Biol.Chem., 280, 2005
|
|
1YM4
 
 | | Crystal structure of human beta secretase complexed with NVP-AMK640 | | Descriptor: | Beta-secretase 1, NVP-AMK640 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
1YM5
 
 | | Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily. | | Descriptor: | Hypothetical 32.6 kDa protein in DAP2-SLT2 intergenic region | | Authors: | Liger, D, Quevillon-Cheruel, S, Sorel, I, Bremang, M, Blondeau, K, Aboulfath, I, Janin, J, Van Tilbeurgh, H, Leulliot, N, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2005-01-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily
Proteins, 60, 2005
|
|
1YM7
 
 | | G Protein-Coupled Receptor Kinase 2 (GRK2) | | Descriptor: | Beta-adrenergic receptor kinase 1 | | Authors: | Lodowski, D.T, Barnhill, J.F, Pyskadlo, R.M, Ghirlando, R, Sterne-Marr, R, Tesmer, J.J.G. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The role of Gbetagamma and domain interfaces in the activation of G protein-coupled receptor kinase 2
Biochemistry, 44, 2005
|
|
1YM8
 
 | |
1YM9
 
 | | Crystal structure of the CDC25B phosphatase catalytic domain with the active site cysteine in the sulfinic form | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2 | | Authors: | Buhrman, G.K, Parker, B, Sohn, J, Rudolph, J, Mattos, C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Mechanism of Oxidative Regulation of the Phosphatase Cdc25B via an Intramolecular Disulfide Bond
Biochemistry, 44, 2005
|
|
1YMA
 
 | |
1YMB
 
 | |
1YMC
 
 | |
1YMD
 
 | | Crystal Structure of the CDC25B phosphatase catalytic domain with the active site cysteine in the sulfonic form | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2 | | Authors: | Buhrman, G.K, Parker, B, Sohn, J, Rudolph, J, Mattos, C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Mechanism of Oxidative Regulation of the Phosphatase Cdc25B via an Intramolecular Disulfide Bond
Biochemistry, 44, 2005
|
|
1YME
 
 | | STRUCTURE OF CARBOXYPEPTIDASE | | Descriptor: | CARBOXYPEPTIDASE A ALPHA, ZINC ION | | Authors: | Greenblatt, H.M, Tucker, P.A, Shoham, G. | | Deposit date: | 1996-07-15 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1YMG
 
 | | The Channel Architecture of Aquaporin O at 2.2 Angstrom Resolution | | Descriptor: | Lens fiber major intrinsic protein, nonyl beta-D-glucopyranoside | | Authors: | Harries, W.E.C, Akhavan, D, Miercke, L.J.W, Khademi, S, Stroud, R.M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Channel Architecture of Aquaporin 0 at a 2.2-A Resolution
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1YMH
 
 | | anti-HCV Fab 19D9D6 complexed with protein L (PpL) mutant A66W | | Descriptor: | Fab 16D9D6, heavy chain, light chain, ... | | Authors: | Granata, V, Housden, N.G, Harrison, S, Jolivet-Reynaud, C, Gore, M.G, Stura, E.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of the crystallization and crystal packing of two Fab single-site mutant protein L complexes.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YMK
 
 | | Crystal Structure of the CDC25B phosphatase catalytic domain in the apo form | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2 | | Authors: | Buhrman, G.K, Parker, B, Sohn, J, Rudolph, J, Mattos, C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Mechanism of Oxidative Regulation of the Phosphatase Cdc25B via an Intramolecular Disulfide Bond
Biochemistry, 44, 2005
|
|
